3VUQ
 
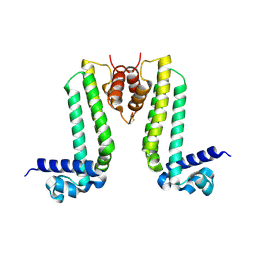 | | Crystal structure of TTHA0167, a transcriptional regulator, TetR/AcrR family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator (TetR/AcrR family) | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-07-04 | | Release date: | 2013-02-27 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and function of a TetR family transcriptional regulator, SbtR, from thermus thermophilus HB8
Proteins, 81, 2013
|
|
3VG8
 
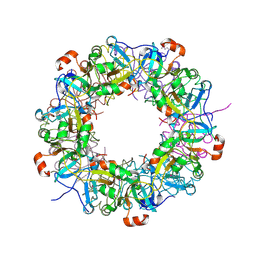 | | Crystal structure of hypothetical protein TTHB210 from Thermus thermophilus HB8 | | Descriptor: | Hypothetical Protein TTHB210 | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein TTHB210, controlled by the [sigma]E/anti-[sigma]E regulatory system in Thermus thermophilus HB8, reveals a novel homodecamer
Proteins, 80, 2012
|
|
3VPR
 
 | | Crystal Structure of a TetR Family Transcriptional Regulator PfmR from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Agari, Y, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2012-03-12 | | Release date: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Transcriptional repression mediated by a TetR family protein, PfmR, from Thermus thermophilus HB8
J.Bacteriol., 2012
|
|
2GBW
 
 | | Crystal Structure of Biphenyl 2,3-Dioxygenase from Sphingomonas yanoikuyae B1 | | Descriptor: | Biphenyl 2,3-Dioxygenase Alpha Subunit, Biphenyl 2,3-Dioxygenase Beta Subunit, FE (III) ION, ... | | Authors: | Ferraro, D.J, Brown, E.N, Yu, C, Parales, R.E, Gibson, D.T, Ramaswamy, S. | | Deposit date: | 2006-03-12 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
Bmc Struct.Biol., 7, 2007
|
|
2GH1
 
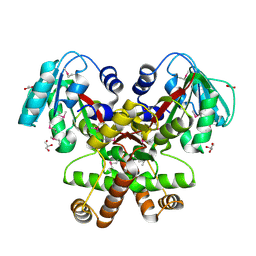 | | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20. | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase | | Authors: | Forouhar, F, Neely, H, Jayaraman, S, Ciao, M, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-24 | | Release date: | 2006-04-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the putative SAM-dependent methyltransferase BC2162 from Bacillus cereus, Northeast Structural Genomics Target BcR20.
To be Published
|
|
2G5V
 
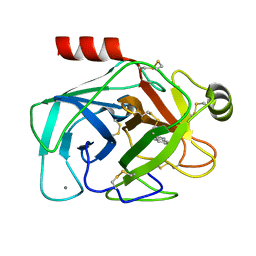 | |
2GBR
 
 | | Crystal Structure of the 35-36 MoaD Insertion Mutant of Ubiquitin | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
2GBX
 
 | | Crystal Structure of Biphenyl 2,3-Dioxygenase from Sphingomonas yanoikuyae B1 Bound to Biphenyl | | Descriptor: | BIPHENYL, Biphenyl 2,3-Dioxygenase Alpha Subunit, Biphenyl 2,3-Dioxygenase Beta Subunit, ... | | Authors: | Ferraro, D.J, Brown, E.N, Yu, C, Parales, R.E, Gibson, D.T, Ramaswamy, S. | | Deposit date: | 2006-03-12 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
Bmc Struct.Biol., 7, 2007
|
|
2GF4
 
 | | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14 | | Descriptor: | ACETATE ION, CALCIUM ION, Protein Vng1086c | | Authors: | Benach, J, Zhou, W, Jayaraman, S, Forouhar, F.F, Janjua, H, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Vng1086c from Halobacterium salinarium (Halobacterium halobium). Northeast Structural Genomics Target HsR14
To be Published
|
|
2GAN
 
 | | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32. | | Descriptor: | 1,2-ETHANEDIOL, 182aa long hypothetical protein, SULFATE ION | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32.
To be Published
|
|
2G5N
 
 | |
2G9D
 
 | | Crystal Structure of Succinylglutamate desuccinylase from Vibrio cholerae, Northeast Structural Genomics Target VcR20 | | Descriptor: | Succinylglutamate desuccinylase | | Authors: | Zhou, W, Jayaraman, S, Forouhar, F, Conover, K, Rong, X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-04-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Succinylglutamate desuccinylase from Vibrio cholerae, Northeast Structural Genomics Target VcR20.
To be Published
|
|
2GBJ
 
 | |
2GBN
 
 | |
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
2GBK
 
 | |
2GGS
 
 | | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii | | Descriptor: | 273aa long hypothetical dTDP-4-dehydrorhamnose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rajakannan, V, Mizushima, T, Suzuki, A, Masui, R, Kuramitsu, S, Yamane, T. | | Deposit date: | 2006-03-24 | | Release date: | 2007-03-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of hypothetical dTDP-4-dehydrorhamnose reductase from sulfolobus tokodaii
To be published
|
|
2GSV
 
 | | X-Ray Crystal Structure of Protein YvfG from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR478. | | Descriptor: | Hypothetical protein yvfG, SULFATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Wang, D, Fang, Y, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Hypothetical Protein YvfG from Bacillus subtilis, Northeast Structural Genomics Target SR478
To be Published
|
|
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8VKP
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKK
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKM
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKO
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
8VKN
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
