4HUJ
 
 | | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti | | Descriptor: | Uncharacterized protein | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti
To be Published
|
|
3B0X
 
 | | K263A mutant of PolX from Thermus thermophilus HB8 complexed with Ca-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakane, S, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-17 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The structural basis of the kinetic mechanism of a gap-filling X-family DNA polymerase that binds Mg(2+)-dNTP before binding to DNA.
J.Mol.Biol., 417, 2012
|
|
2QYH
 
 | | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426 | | Descriptor: | GLYCEROL, Hypothetical conserved protein, GK1056 | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-08-15 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hypothetical protein (gk1056) from geobacillus kaustophilus HTA426
To be Published
|
|
2D61
 
 | | Aspartate Aminotransferase Mutant MA With Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D63
 
 | | Aspartate Aminotransferase Mutant MA With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D5Y
 
 | | Aspartate Aminotransferase Mutant MC With Isovaleric Acid | | Descriptor: | Aspartate aminotransferase, ISOVALERIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D7Y
 
 | | Aspartate Aminotransferase Mutant MA | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D3Y
 
 | | Crystal structure of uracil-DNA glycosylase from Thermus Thermophilus HB8 | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ACETATE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2D66
 
 | | Aspartate Aminotransferase Mutant MAB | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2QQ3
 
 | | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-CoA hydratase subunit I | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Of Enoyl-CoA Hydrates Subunit I (gk_2039) Other Form From Geobacillus Kaustophilus HTA426
To be Published
|
|
2D4R
 
 | | Crystal structure of TTHA0849 from Thermus thermophilus HB8 | | Descriptor: | SULFATE ION, hypothetical protein TTHA0849 | | Authors: | Nakabayashi, M, Shibata, N, Kuramitsu, S, Higuchi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: a putative member of the StAR-related lipid-transfer (START) domain superfamily.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2YVS
 
 | |
2D7Z
 
 | | Aspartate Aminotransferase Mutant MAB Complexed with Maleic Acid | | Descriptor: | Aspartate aminotransferase, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-30 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2D65
 
 | | Aspartate Aminotransferase Mutant MABC | | Descriptor: | Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tanaka, Y, Nakagawa, N, Tada, H, Yano, T, Masui, R, Kuramitsu, S. | | Deposit date: | 2005-11-09 | | Release date: | 2006-11-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structures of Aspartate Aminotransferase with Mutations of Non-Active-Site Residues
To be Published
|
|
2QVG
 
 | |
4G9Q
 
 | | Crystal structure of a 4-carboxymuconolactone decarboxylase | | Descriptor: | 4-carboxymuconolactone decarboxylase | | Authors: | Hickey, H.D, Mcgillick, B.E, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-24 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a 4-carboxymuconolactone decarboxylase
To be Published
|
|
2RBG
 
 | |
2DDG
 
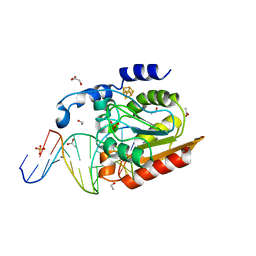 | | Crystal structure of uracil-DNA glycosylase in complex with AP:G containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*GP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-01-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2DP6
 
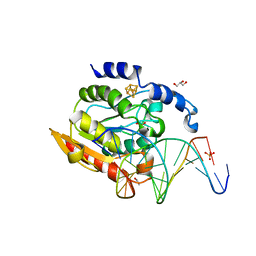 | | Crystal structure of uracil-DNA glycosylase in complex with AP:C containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*CP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Kuramitsu, S, Hoseki, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-07 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Family 5 Uracil-DNA Glycosylase Bound to DNA Reveals Insights into the Mechanism for Substrate Recognition and Catalysis
To be Published
|
|
2ZKI
 
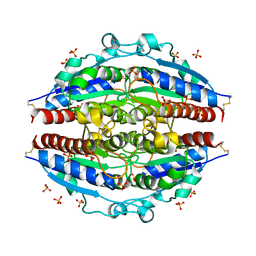 | | Crystal structure of hypothetical Trp repressor binding protein from Sul folobus tokodaii (ST0872) | | Descriptor: | 199aa long hypothetical Trp repressor binding protein, SULFATE ION | | Authors: | Kawano, T, Teshima, N, Suzuki, A, Kuramitsu, S, Yamane, T. | | Deposit date: | 2008-03-21 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of hypothetical Trp repressor binding protein from Sul
folobus tokodaii (ST0872)
To be Published
|
|
2RK9
 
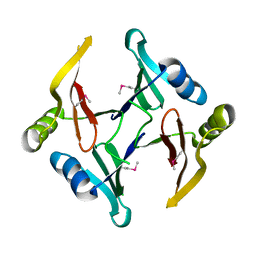 | | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01 | | Descriptor: | Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Tyagi, R, Eswaramoorthy, S, Sauder, J.M, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein/dioxygenase superfamily member from Vibrio splendidus 12B01.
To be Published
|
|
2DEM
 
 | | Crystal structure of Uracil-DNA glycosylase in complex with AP:A containing DNA | | Descriptor: | 5'-D(*AP*TP*GP*TP*TP*GP*CP*(D1P)P*TP*TP*AP*GP*TP*CP*C)-3', 5'-D(*GP*GP*AP*CP*TP*AP*AP*AP*GP*CP*AP*AP*CP*A)-3', DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kosaka, H, Nakagawa, N, Masui, R, Hoseki, J, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-13 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of family 5 uracil-DNA glycosylase bound to DNA.
J.Mol.Biol., 373, 2007
|
|
2ZXO
 
 | | Crystal structure of RecJ from Thermus thermophilus HB8 | | Descriptor: | Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
2ZXP
 
 | | Crystal structure of RecJ in complex with Mn2+ from Thermus thermophilus HB8 | | Descriptor: | MANGANESE (II) ION, Single-stranded DNA specific exonuclease RecJ | | Authors: | Wakamatsu, T, Kitamura, Y, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of RecJ exonuclease defines its specificity for single-stranded DNA
J.Biol.Chem., 285, 2010
|
|
2EC2
 
 | | Crystal structure of transposase from Sulfolobus tokodaii | | Descriptor: | 136aa long hypothetical transposase, SULFATE ION | | Authors: | Kawai, K, Suzuki, A, Kuramitsu, S, Masui, R, Yamane, T. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of transposase from Sulfolobus tokodaii
To be Published
|
|
