7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWS
 
 | | Crystal structure of tamarin CD81 large extracellular loop | | Descriptor: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
3FY7
 
 | | Crystal structure of homo sapiens CLIC3 | | Descriptor: | Chloride intracellular channel protein 3, SULFATE ION | | Authors: | Littler, D.R, Curmi, P.M.G, Breit, S.N, Perrakis, A. | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of human CLIC3 at 2 A resolution
Proteins, 78, 2010
|
|
3O1X
 
 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C84A mutant from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
7RCK
 
 | | Crystal Structure of PMS2 with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7RCI
 
 | | Crystal Structure of a PMS2 VUS with Substrate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mismatch repair endonuclease PMS2 | | Authors: | D'Arcy, B.M, Prakash, A. | | Deposit date: | 2021-07-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | PMS2 variant results in loss of ATPase activity without compromising mismatch repair.
Mol Genet Genomic Med, 10, 2022
|
|
7A5T
 
 | | Crystal structure of A55E mutant of BlaC from Mycobacterium tuberculosis | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-21 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7A71
 
 | | Structure of G132S BlaC from Mycobacterium tuberculosis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The G132S Mutation Enhances the Resistance of Mycobacterium tuberculosis beta-Lactamase against Sulbactam.
Biochemistry, 60, 2021
|
|
7Z5G
 
 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6S
 
 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
5D75
 
 | | Crystal structure of Human FKBD25 in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Peptidyl-prolyl cis-trans isomerase FKBP3 | | Authors: | Rajan, S, Prakash, A, Yoon, H.S. | | Deposit date: | 2015-08-13 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the FK506 binding domain of human FKBP25 in complex with FK506.
Protein Sci., 25, 2016
|
|
7ZW4
 
 | | Crystal structure of Talin R7R8 domains with Caskin-2 LD-peptide | | Descriptor: | Caskin-2, Talin-1 | | Authors: | Celie, P.H.N, Joosten, R.P, Sonnenberg, A, Perrakis, A. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Caskin2 is a novel talin- and Abi1-binding protein that promotes cell motility.
J.Cell.Sci., 137, 2024
|
|
3QGZ
 
 | | Re-investigated high resolution crystal structure of histidine triad nucleotide-binding protein 1 (HINT1) from rabbit complexed with adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A, Nawrot, B, Stec, W.J. | | Deposit date: | 2011-01-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution X-ray crystal structure of rabbit histidine triad nucleotide-binding protein 1 (rHINT1) - adenosine complex at 1.10A resolution
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3O1C
 
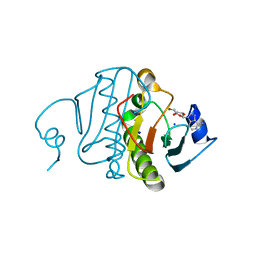 | | High resolution crystal structure of histidine triad nucleotide-binding protein 1 (Hint1) C38A mutant from rabbit complexed with Adenosine | | Descriptor: | ADENOSINE, Histidine triad nucleotide-binding protein 1, SODIUM ION | | Authors: | Dolot, R.M, Ozga, M, Krakowiak, A.K, Nawrot, B, Stec, W.J. | | Deposit date: | 2010-07-21 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Histidine Triad Nucleotide-binding Protein 1 (HINT-1) Phosphoramidase Transforms Nucleoside 5'-O-Phosphorothioates to Nucleoside 5'-O-Phosphates.
J.Biol.Chem., 285, 2010
|
|
4G91
 
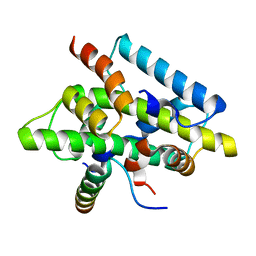 | | CCAAT-binding complex from Aspergillus nidulans | | Descriptor: | HAPB protein, HapE, Transcription factor HapC (Eurofung) | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
7Z3L
 
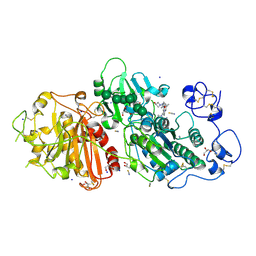 | | Autotaxin in complex with hybrid compound ziritaxestat (GLPG1690) | | Descriptor: | 2-[[2-ethyl-8-methyl-6-[4-[2-(3-oxidanylazetidin-1-yl)-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Ford, P, Heckmann, B, Perrakis, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autotaxin facilitates selective LPA receptor signaling.
Cell Chem Biol, 30, 2023
|
|
7ZOT
 
 | |
1NAW
 
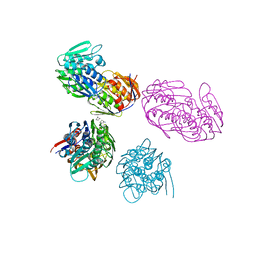 | | ENOLPYRUVYL TRANSFERASE | | Descriptor: | CYCLOHEXYLAMMONIUM ION, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYL-TRANSFERASE | | Authors: | Schoenbrunn, E, Sack, S, Eschenburg, S, Perrakis, A, Krekel, F, Amrhein, N, Mandelkow, E. | | Deposit date: | 1996-07-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine enolpyruvyltransferase, the target of the antibiotic fosfomycin.
Structure, 4, 1996
|
|
2V0S
 
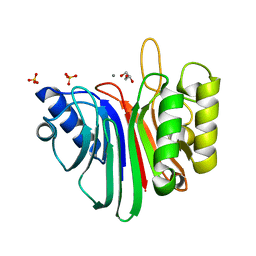 | | crystal structure of a hairpin exchange variant (LR1) of the targeting LINE-1 retrotransposon endonuclease | | Descriptor: | GLYCEROL, LR1, MANGANESE (II) ION, ... | | Authors: | Repanas, K, Zingler, N, Layer, L.E, Schumann, G.G, Perrakis, A, Weichenrieder, O. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determinants for DNA Target Structure Selectivity of the Human Line-1 Retrotransposon Endonuclease.
Nucleic Acids Res., 35, 2007
|
|
4G92
 
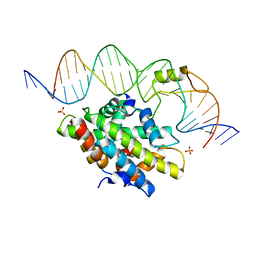 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | Descriptor: | DNA, HAPB protein, HapE, ... | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
2L99
 
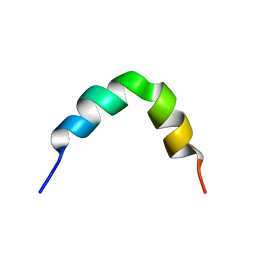 | | Solution structure of LAK160-P10 | | Descriptor: | LAK160-P10 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L9A
 
 | | Solution structure of LAK160-P12 | | Descriptor: | LAK160-P12 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
7Z0N
 
 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
5CE8
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis | | Descriptor: | Branched-chain amino acid aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for L-amino acids and R-amines.
Extremophiles, 20, 2016
|
|
