3EGG
 
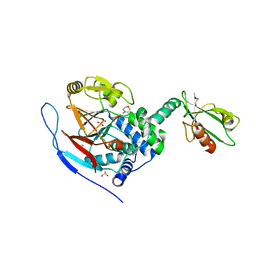 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EGH
 
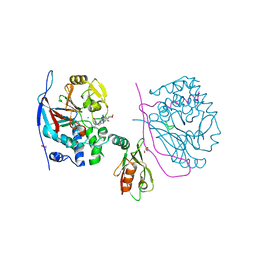 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6NNB
 
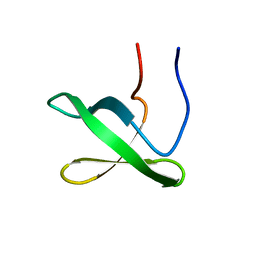 | |
5WLP
 
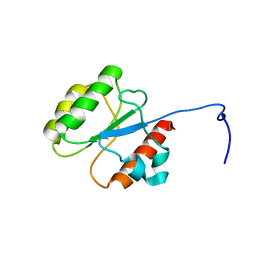 | |
8UF3
 
 | |
6Q2U
 
 | | Structure of Cytochrome C4 from Pseudomonas aeruginosa | | Descriptor: | Cytochrome c4, HEME C | | Authors: | Carpenter, J.M, Zhong, F, Pletneva, E.V, Ragusa, M.J. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and redox properties of the diheme electron carrier cytochrome c4from Pseudomonas aeruginosa.
J.Inorg.Biochem., 203, 2019
|
|
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4HPQ
 
 | | Crystal Structure of the Atg17-Atg31-Atg29 Complex | | Descriptor: | Atg17, Atg29, Atg31 | | Authors: | Stanley, R.E, Ragusa, M.J, Hurley, J.H. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Architecture of the atg17 complex as a scaffold for autophagosome biogenesis.
Cell(Cambridge,Mass.), 151, 2012
|
|
4J2G
 
 | | Atg13 HORMA domain | | Descriptor: | KLTH0A00704p, SULFATE ION | | Authors: | Jao, C, Stanley, R.E, Ragusa, M.J, Hurley, J.H. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6VZF
 
 | | Crystal Structure of Atg11 Coiled-Coil 3 | | Descriptor: | Autophagy-related protein 11, SULFATE ION | | Authors: | Margolis, H.K, Ragusa, M.J. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Third Coiled Coil Domain of Atg11 Is Required for Shaping Mitophagy Initiation Sites.
J.Mol.Biol., 432, 2020
|
|
6W9N
 
 | |
6MI3
 
 | | Structure of NEMO(51-112) with N- and C-terminal coiled-coil adaptors. | | Descriptor: | NF-kB ESSENTIAL MODULATOR,NF-kappa-B essential modulator,NF-kB ESSENTIAL MODULATOR | | Authors: | Pellegrini, M, Barczewski, A.H, Mierke, D.F, Ragusa, M.J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface.
Sci Rep, 9, 2019
|
|
6MI4
 
 | |
