1PCK
 
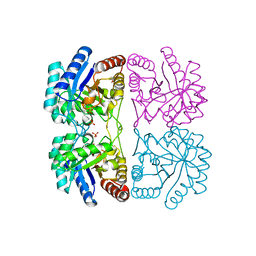 | | Aquifex aeolicus KDO8PS in complex with Z-methyl-PEP | | Descriptor: | 2-(PHOSPHONOOXY)BUTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Wang, J, Xu, X, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
DRUG DES.DISCOVERY, 18, 2003
|
|
1PBL
 
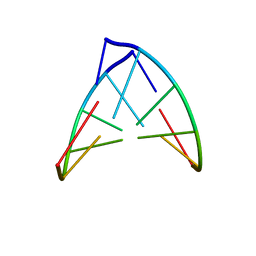 | | STRUCTURE OF RIBONUCLEIC ACID, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*OMCP*OMGP*OMCP*OMGP*OMCP*OMG)-3') | | Authors: | Popenda, M, Biala, E, Milecki, J, Adamiak, R.W. | | Deposit date: | 1996-08-05 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2'-O-Me(CGCGCG)2 under low salt conditions.
Nucleic Acids Res., 25, 1997
|
|
1PCI
 
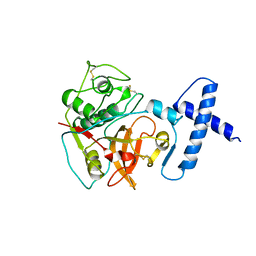 | | PROCARICAIN | | Descriptor: | PROCARICAIN | | Authors: | Groves, M.R, Taylor, M.A.J, Scott, M, Cummings, N.J, Pickersgill, R.W, Jenkins, J.A. | | Deposit date: | 1996-06-28 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft.
Structure, 4, 1996
|
|
1PCW
 
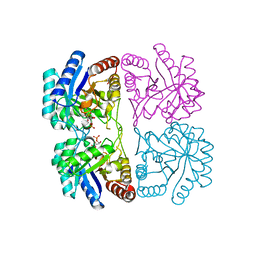 | | Aquifex aeolicus KDO8PS in complex with cadmium and APP, a bisubstrate inhibitor | | Descriptor: | 1-DEOXY-6-O-PHOSPHONO-1-[(PHOSPHONOMETHYL)AMINO]-L-THREO-HEXITOL, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Xu, X, Wang, J, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-17 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
Drug DES.DISCOVERY, 18, 2003
|
|
1T3E
 
 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
1RU4
 
 | | Crystal structure of pectate lyase Pel9A | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Jenkins, J, Shevchik, V.E, Hugouvieux-Cotte-Pattat, N, Pickersgill, R.W. | | Deposit date: | 2003-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi
J.Biol.Chem., 279, 2004
|
|
1PBM
 
 | | STRUCTURE OF RIBONUCLEIC ACID, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Popenda, M, Biala, E, Milecki, J, Adamiak, R.W. | | Deposit date: | 1996-08-05 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2'-O-Me(CGCGCG)2 under low salt conditions.
Nucleic Acids Res., 25, 1997
|
|
1PPO
 
 | | DETERMINATION OF THE STRUCTURE OF PAPAYA PROTEASE OMEGA | | Descriptor: | MERCURY (II) ION, PROTEASE OMEGA | | Authors: | Pickersgill, R.W, Rizkallah, P.J, Harris, G.W, Goodenough, P.W. | | Deposit date: | 1991-07-12 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Determination of the Structure of Papaya Protease Omega
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1T4X
 
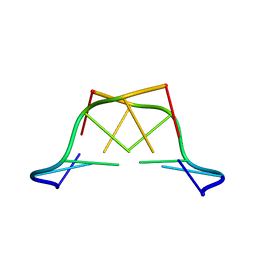 | | The first left-handed RNA structure of (CGCGCG)2, Z-RNA, NMR, 12 structures, determined in high salt | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Popenda, M, Milecki, J, Adamiak, R.W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High salt solution structure of a left-handed RNA double helix.
Nucleic Acids Res., 32, 2004
|
|
1TNH
 
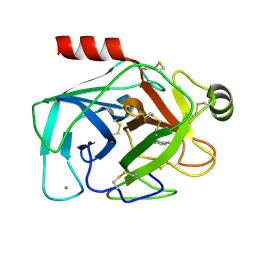 | |
1TNJ
 
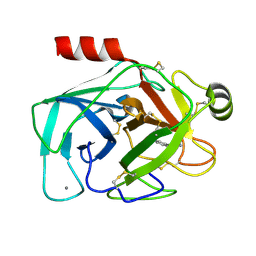 | |
1TNG
 
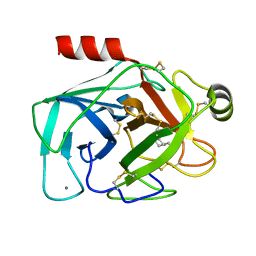 | |
2AOC
 
 | | Crystal structure analysis of HIV-1 protease mutant I84V with a substrate analog P2-NC | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2B1V
 
 | | Human estrogen receptor alpha ligand-binding domain in complex with OBCP-1M and a glucocorticoid receptor interacting protein 1 NR box II peptide | | Descriptor: | 4-[(1S,2S,5S)-5-(HYDROXYMETHYL)-8-METHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
2AOG
 
 | | Crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog P2-NC | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 PROTEASE (RETROPEPSIN), ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2C56
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | URACIL DNA GLYCOSYLASE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Krusong, K, Carpenter, E.P, Bellamy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2BO0
 
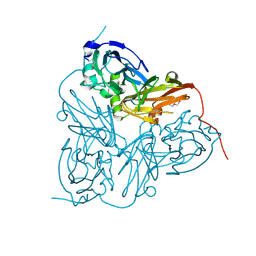 | | Crystal structure of the C130A mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2C53
 
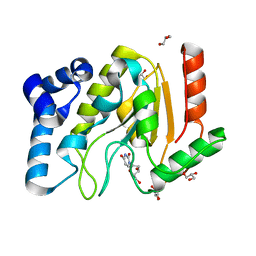 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | Authors: | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2BWI
 
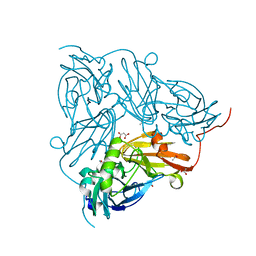 | | Atomic Resolution Structure of Nitrite -soaked Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BP0
 
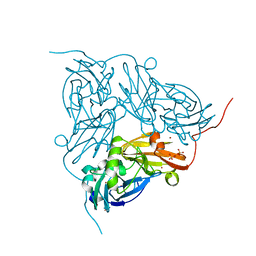 | | M144L mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, SULFATE ION, ... | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-17 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2BWD
 
 | | Atomic Resolution Structure of Achromobacter cycloclastes Cu Nitrite Reductase with Endogenously bound Nitrite and NO | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-13 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BRT
 
 | | ANTHOCYANIDIN SYNTHASE FROM ARABIDOPSIS THALIANA COMPLEXED with naringenin | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, LEUCOANTHOCYANIDIN DIOXYGENASE, ... | | Authors: | Turnbull, J.J, Clifton, I.J, Welford, R.W.D, Schofield, C.J. | | Deposit date: | 2005-05-11 | | Release date: | 2006-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Studies on Anthocyanidin Synthase Catalysed Oxidation of Flavanone Substrates: The Effect of C-2 Stereochemistry on Product Selectivity and Mechanism
Org.Biomol.Chem., 3, 2005
|
|
2BW4
 
 | | Atomic Resolution Structure of Resting State of the Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BW5
 
 | | Atomic Resolution Structure of NO-bound Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-12 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
