3OF9
 
 | |
4I63
 
 | | Crystal Structure of E-R ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.709 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4ERJ
 
 | |
4HW0
 
 | | Crystal structure of Sso10a-2, a DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | DNA-binding protein Sso10a-2 | | Authors: | Waterreus, W.J, Goosen, N, Moolenaar, G.F, Driessen, R.P.C, Dame, R.T, Pannu, N.S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-10-30 | | Last modified: | 2017-01-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse architectural properties of Sso10a proteins: Evidence for a role in chromatin compaction and organization.
Sci Rep, 6, 2016
|
|
4FEL
 
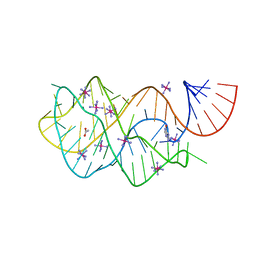 | | Crystal structure of the U25A/A46G mutant of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
3OU0
 
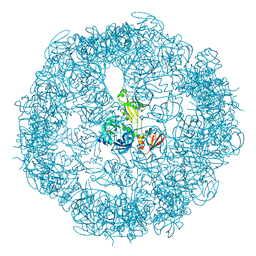 | | re-refined 3CS0 | | Descriptor: | Periplasmic serine endoprotease DegP, heptapeptide, pentapeptide | | Authors: | Sauer, R.T, Grant, R.A, Kim, S. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Cell(Cambridge,Mass.), 145, 2011
|
|
4FEP
 
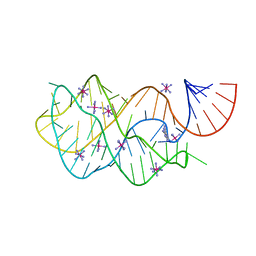 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4ISZ
 
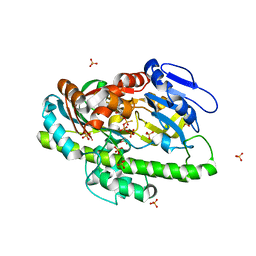 | | RNA ligase RtcB in complex with GTP alphaS and Mn(II) | | Descriptor: | GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IWN
 
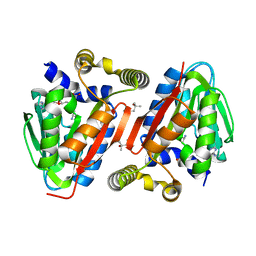 | | Crystal structure of a putative methyltransferase CmoA in complex with a novel SAM derivative | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, tRNA (cmo5U34)-methyltransferase | | Authors: | Aller, P, Lobley, C.M, Byrne, R.T, Antson, A.A, Waterman, D.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | S-Adenosyl-S-carboxymethyl-L-homocysteine: a novel cofactor found in the putative tRNA-modifying enzyme CmoA.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NLA
 
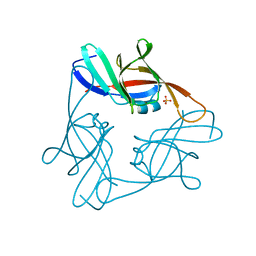 | | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein, apo form | | Descriptor: | Iron-regulated surface determinant protein A, SULFATE ION | | Authors: | Malmirchegini, G.R, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2013-11-14 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes.
J.Biol.Chem., 289, 2014
|
|
4NEU
 
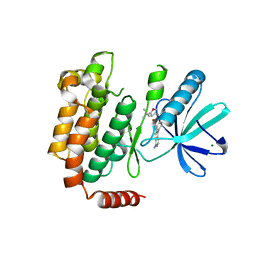 | | X-ray structure of Receptor Interacting Protein 1 (RIP1)kinase domain with a 1-aminoisoquinoline inhibitor | | Descriptor: | 1-[4-(1-aminoisoquinolin-5-yl)phenyl]-3-(5-tert-butyl-1,2-oxazol-3-yl)urea, MAGNESIUM ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Nolte, R.T, Ward, P, kahler, K.M, Campobasso, N. | | Deposit date: | 2013-10-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis.
ACS Med Chem Lett, 4, 2013
|
|
4IXL
 
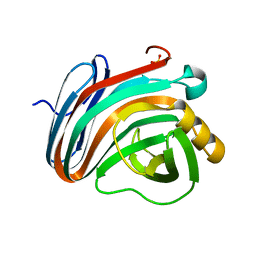 | | Crystal structure of endo-beta-1,4-xylanase from the alkaliphilic Bacillus sp. SN5 | | Descriptor: | SULFATE ION, xylanase | | Authors: | Bai, W.Q, Xue, Y.F, Huang, J.X, Zhou, C, Guo, R.T, Ma, Y.H. | | Deposit date: | 2013-01-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure Analysis of Alkaline xylanase from Alkaliphilic Bacillus sp. SN5 Implications for pH-dependent enzyme activity
To be Published
|
|
4FEO
 
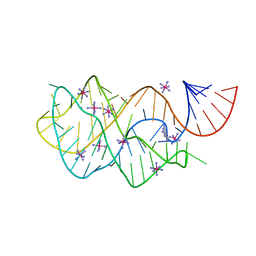 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FE5
 
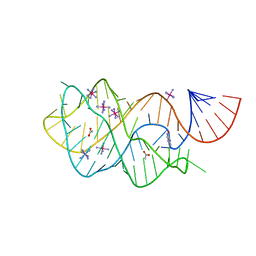 | | Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine.
Nature, 432, 2004
|
|
4N2P
 
 | | Structure of Archease from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-10-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.435 Å) | | Cite: | A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation.
Nucleic Acids Res., 42, 2014
|
|
4FEN
 
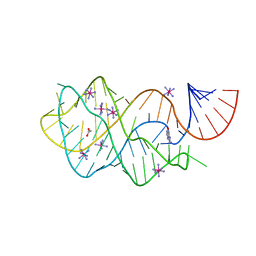 | | Crystal structure of the A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U/U25A/A46G mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FEJ
 
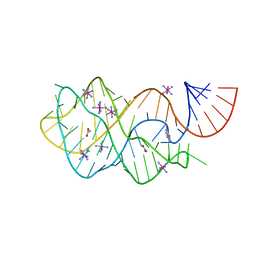 | | Crystal structure of the A24U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4GMA
 
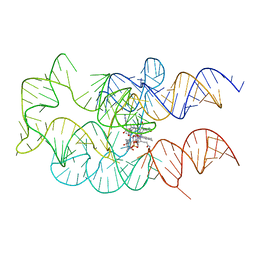 | | Crystal structure of the adenosylcobalamin riboswitch | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4PER
 
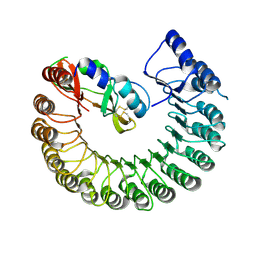 | | Structure of Gallus gallus ribonuclease inhibitor complexed with Gallus gallus ribonuclease I | | Descriptor: | Angiogenin, Ribonuclease Inhibitor | | Authors: | Bianchetti, C.M, Lomax, J.E, Raines, R.T, Fox, B.G. | | Deposit date: | 2014-04-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
4PEQ
 
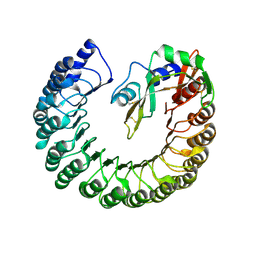 | | Structure of bovine ribonuclease inhibitor complexed with bovine ribonuclease I | | Descriptor: | Ribonuclease pancreatic, Ribonuclease/angiogenin inhibitor 1 | | Authors: | Bianchetti, C.M, Lomax, J.E, Raines, R.T, Fox, B.G. | | Deposit date: | 2014-04-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Functional evolution of ribonuclease inhibitor: insights from birds and reptiles.
J.Mol.Biol., 426, 2014
|
|
4KTE
 
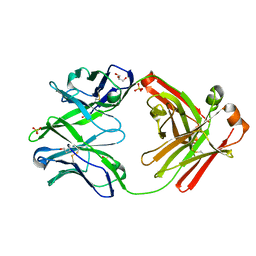 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3RSD
 
 | |
3RSK
 
 | |
4ISJ
 
 | | RNA Ligase RtcB in complex with Mn(II) | | Descriptor: | MANGANESE (II) ION, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.344 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IT0
 
 | | Structure of the RNA ligase RtcB-GMP/Mn(II) complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
