7WGH
 
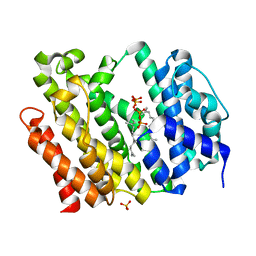 | | Crystal structure of AflSQS from Aspergillus flavus in complex with FSPP | | Descriptor: | PHOSPHATE ION, PYROPHOSPHATE 2-, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Shang, N, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Structural and Bioinformatics Investigation of a Fungal Squalene Synthase and Comparisons with Other Membrane Proteins.
Acs Omega, 7, 2022
|
|
7Y8L
 
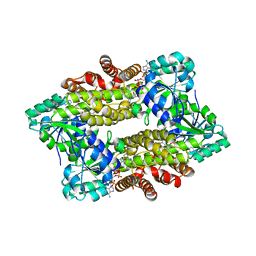 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7VQ6
 
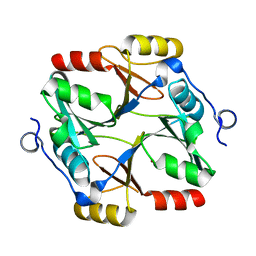 | | Structure of a specialized glyoxalase from Gossypium hirsutum | | Descriptor: | Lactoylglutathione lyase, NICKEL (II) ION | | Authors: | Li, H, Hu, Y.M, Dai, L.H, Chen, C.C, Huang, J.W, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure and biochemical analysis of the specialized deoxynivalenol-detoxifying glyoxalase SPG from Gossypium hirsutum.
Int.J.Biol.Macromol., 200, 2022
|
|
7Y8K
 
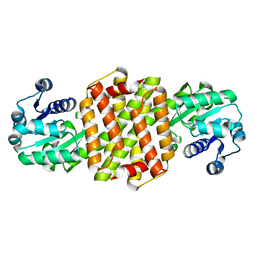 | | Structure of ScIRED wild-type from Streptomyces clavuligerus | | Descriptor: | Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
2WCY
 
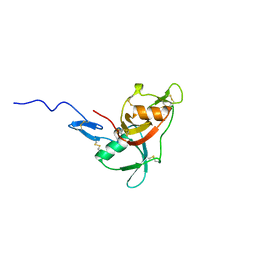 | | NMR solution structure of factor I-like modules of complement C7. | | Descriptor: | COMPLEMENT COMPONENT C7 | | Authors: | Phelan, M.M, Thai, C.T, Soares, D.C, Ogata, R.T, Barlow, P.N, Bramham, J. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Factor I-Like Modules from Complement C7 Reveals a Pair of Follistatin Domains in Compact Pseudosymmetric Arrangement.
J.Biol.Chem., 284, 2009
|
|
7XWM
 
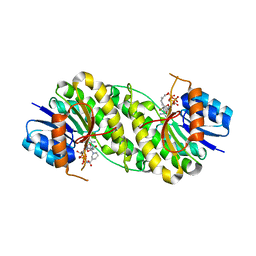 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7Y8N
 
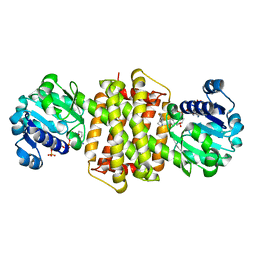 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7XWJ
 
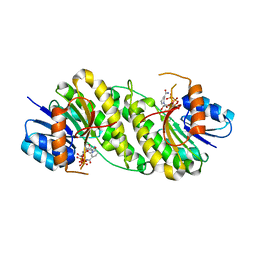 | | structure of patulin-detoxifying enzyme Y155F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWI
 
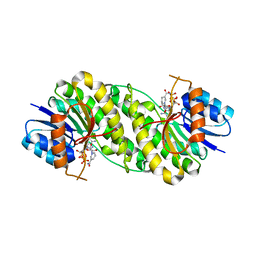 | | structure of patulin-detoxifying enzyme with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWH
 
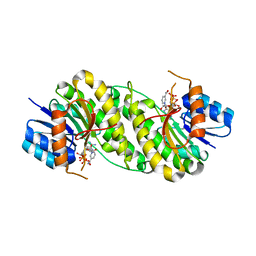 | | structure of patulin-detoxifying enzyme with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWL
 
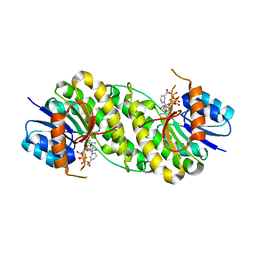 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWN
 
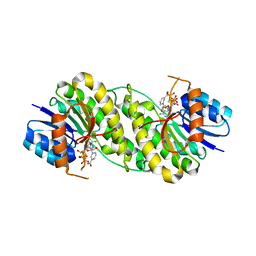 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
5XK7
 
 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with DMAPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XO6
 
 | | Crystal structure of a novel ZEN lactonase mutant | | Descriptor: | PENTAETHYLENE GLYCOL, lactonase for protein | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-05-27 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of a Mycoestrogen-Detoxifying Lactonase from Rhinocladiella mackenziei: Molecular Insight into ZHD Substrate Selectivity
Acs Catalysis, 8, 2018
|
|
5YGK
 
 | | Crystal structure of a synthase from Streptomyces sp. CL190 with dmaspp | | Descriptor: | Cyclolavandulyl diphosphate synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
5YNW
 
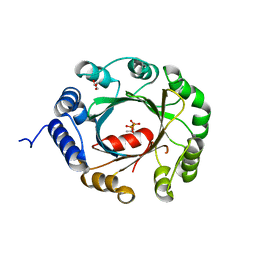 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP and INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YGJ
 
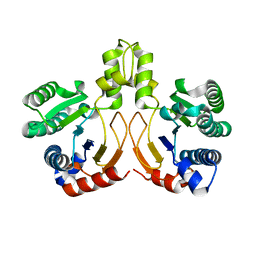 | | Crystal structure of a synthase from Streptomyces sp. CL190 | | Descriptor: | Cyclolavandulyl diphosphate synthase | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
5XZU
 
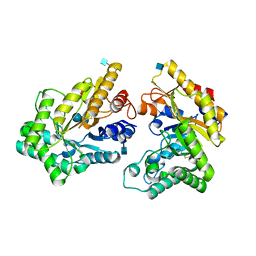 | | Crystal structure of GH10 xylanase from Bispora. sp MEY-1 with xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | You, S, Chen, C.C, Tu, T, Guo, R.T, Luo, H.Y, Yao, B. | | Deposit date: | 2017-07-14 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into the functional roles of Glu175 in the hyperthermostable xylanase XYL10C-Delta N through structural analysis and site-saturation mutagenesis.
Biotechnol Biofuels, 11, 2018
|
|
5YNT
 
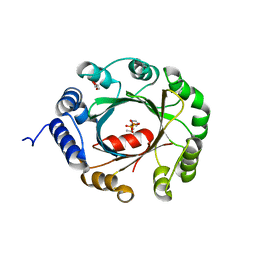 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YNV
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, aromatic prenyltransferase | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
5YNU
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
