4L81
 
 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92, deltaG93)) | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2013-06-15 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KT8
 
 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | Descriptor: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
4I34
 
 | | Crystal Structure of W-W-W ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-23 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1218 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I81
 
 | | Crystal Structure of ATPgS bound ClpX Hexamer | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-12-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8182 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I4L
 
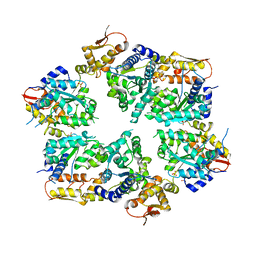 | | Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6981 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4ISJ
 
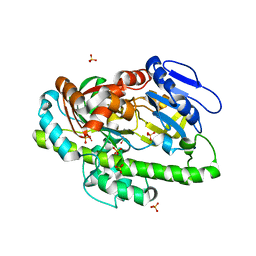 | | RNA Ligase RtcB in complex with Mn(II) | | Descriptor: | MANGANESE (II) ION, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.344 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IT0
 
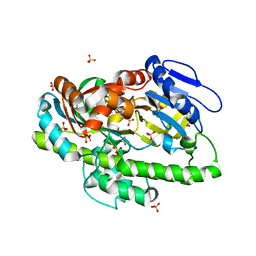 | | Structure of the RNA ligase RtcB-GMP/Mn(II) complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4O89
 
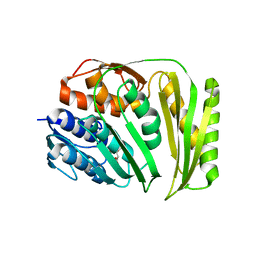 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
4OQU
 
 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92)) | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, SAM-I/IV riboswitch | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OJR
 
 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | Authors: | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | Deposit date: | 2014-01-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
4LFD
 
 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
4KPF
 
 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (3aS,4R)-4-amino-13-cyclopropyl-8-fluoro-10-oxo-3a,4,5,6,10,13-hexahydro-1H,3H-pyrrolo[2',1':3,4][1,4]oxazepino[5,6-h]quinoline-11-carboxylic acid, E-site1, E-site2, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
4MYP
 
 | | Structure of the central NEAT domain, N2, of the listerial Hbp2 protein complexed with heme | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Malmirchegini, G.R, Sawaya, M.R, Clubb, R.T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-binding Hemophore from Listeria monocytogenes.
J.Biol.Chem., 289, 2014
|
|
4LE4
 
 | | Crystal structure of PaGluc131A with cellotriose | | Descriptor: | Beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4LE3
 
 | | Crystal structure of a GH131 beta-glucanase catalytic domain from Podospora anserina | | Descriptor: | Beta-glucanase | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4Q2Z
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE356, from a non-human primate | | Descriptor: | Heavy chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, Light chain of Fab fragment of HIV vaccine-elicited CD4bs-directed antibody | | Authors: | Navis, M, Tran, K, Bale, S, Phad, G, Guenaga, J, Wilson, R, Soldemo, M, McKee, K, Sundling, C, Mascola, J, Li, Y, Wyatt, R.T, Hedestam, G.B.K. | | Deposit date: | 2014-04-10 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization.
Plos Pathog., 10, 2014
|
|
1L7F
 
 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
1LLI
 
 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1L7G
 
 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
6J0L
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with sulfate ion | | Descriptor: | Butyrophilin subfamily 3 member A3, SULFATE ION | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J0K
 
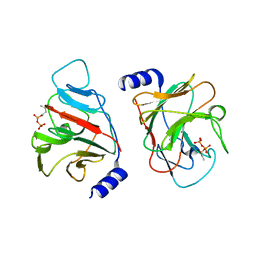 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
1LLW
 
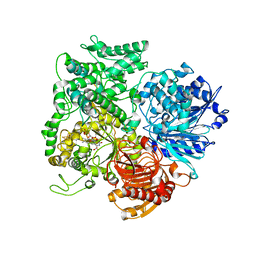 | | Structural studies on the synchronization of catalytic centers in glutamate synthase: complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | van den Heuvel, R.H, Ferrari, D, Bossi, R.T, Ravasio, S, Curti, B, Vanoni, M.A, Florencio, F.J, Mattevi, A. | | Deposit date: | 2002-04-30 | | Release date: | 2002-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies on the synchronization of catalytic centers in glutamate synthase
J.BIOL.CHEM., 277, 2002
|
|
6J0G
 
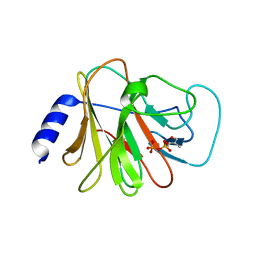 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J03
 
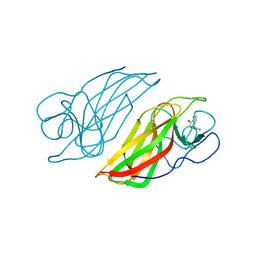 | | Crystal structure of a cyclase mutant in apo form | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Tang, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the calcium dependence of Stig cyclases.
Rsc Adv, 9, 2019
|
|
6J06
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP-08 | | Descriptor: | (2E)-3-(hydroxymethyl)-4-(4-methylphenyl)but-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1, CALCIUM ION, ... | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
