6PP8
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
6PP6
 
 | | ClpX in ClpX-ClpP complex bound to substrate and ATP-gamma-S, class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Fei, X, Jenni, S, Harrison, S.C, Sauer, R.T. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structures of the ATP-fueled ClpXP proteolytic machine bound to protein substrate.
Elife, 9, 2020
|
|
4FZS
 
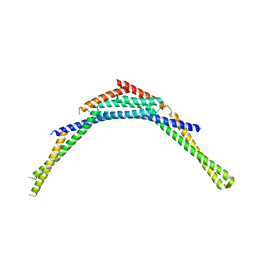 | | Structure of human SNX1 BAR domain | | Descriptor: | Sorting nexin-1 | | Authors: | van Weering, J.R.T, Sessions, R.B, Traer, C.J, Kloer, D.P, Bhatia, V.K, Stamou, D, Hurley, J.H, Cullen, P.J. | | Deposit date: | 2012-07-07 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insight into vesicle-to-tubule membrane remodeling by SNX-BAR proteins
To be Published
|
|
6PXI
 
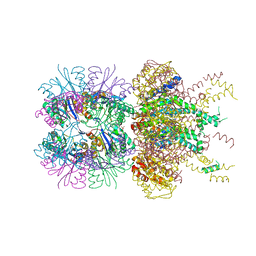 | | The crystal structure of a singly capped HslUV complex with an axial pore plug and a HslU E257Q mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.447 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PVU
 
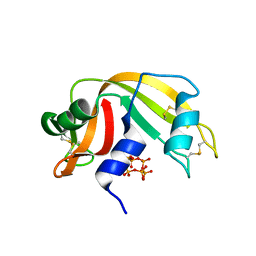 | | RNase A in complex with hexametaphosphate | | Descriptor: | 2,4,6,8,10,12-hexahydroxy-2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~,10lambda~5~,12lambda~5~-cyclohexaphosphoxane-2,4,6,8,10,12-hexone, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PVW
 
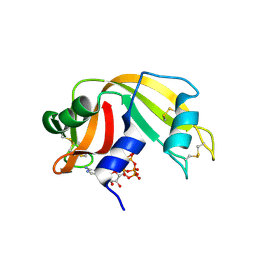 | | RNase A in complex with cp4pA | | Descriptor: | 2,4,6,8-tetrahydroxy-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocane-2,4,6,8-tetrone, 5'-O-[(R)-hydroxy{[(4R,8S)-4,6,8-trihydroxy-2,4,6,8-tetraoxo-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocan-2-yl]oxy}phosphoryl]adenosine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
4KPF
 
 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (3aS,4R)-4-amino-13-cyclopropyl-8-fluoro-10-oxo-3a,4,5,6,10,13-hexahydro-1H,3H-pyrrolo[2',1':3,4][1,4]oxazepino[5,6-h]quinoline-11-carboxylic acid, E-site1, E-site2, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
4L81
 
 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92, deltaG93)) | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2013-06-15 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6PVX
 
 | | RNase A in complex with p5U | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy{[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]oxy}phosphoryl]uridine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PVV
 
 | | RNase A in complex with p5A | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
6PXL
 
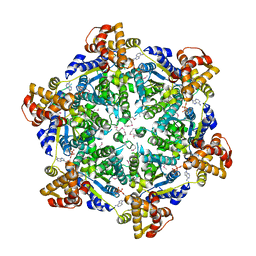 | | 3.74 Angstroms resolution structure of HlsU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, MAGNESIUM ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.741 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
4FE5
 
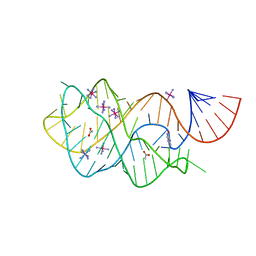 | | Crystal structure of the xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine.
Nature, 432, 2004
|
|
6PXK
 
 | | 3.65 Angstroms resolution structure of HslU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, SULFATE ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.647 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
4FEJ
 
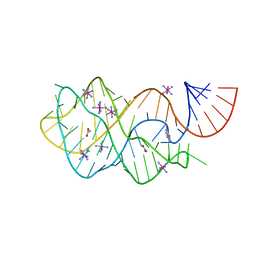 | | Crystal structure of the A24U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4LFD
 
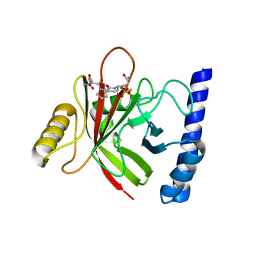 | | Staphylococcus aureus sortase B-substrate complex | | Descriptor: | (CBZ)NPQ(B27) PEPTIDE, SULFATE ION, Sortase B | | Authors: | Jacobitz, A.W, Sawaya, M.R, Yi, S.W, Amer, B.R, Huang, G.L, Nguyen, A.V, Jung, M.E, Clubb, R.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
J.Biol.Chem., 289, 2014
|
|
4FEN
 
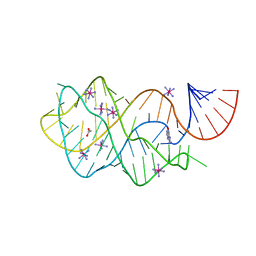 | | Crystal structure of the A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | | Descriptor: | A24U/U25A/A46G mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4FRN
 
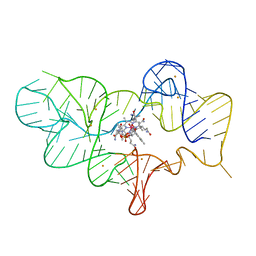 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
6QPX
 
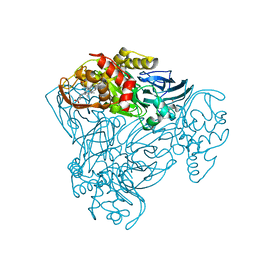 | | Crystal structure of nitrite bound Y323A mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ2
 
 | | Crystal structure of nitrite bound Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ1
 
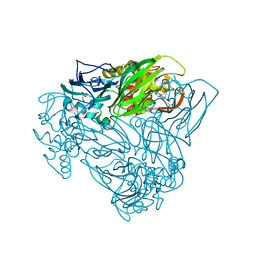 | | Crystal structure of as isolated Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
4KTE
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FEP
 
 | | Crystal structure of the A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, A24U/U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain, COBALT HEXAMMINE(III) | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
4LE4
 
 | | Crystal structure of PaGluc131A with cellotriose | | Descriptor: | Beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, T, Chan, H.C, Huang, C.H, Ko, T.P, Huang, T.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a GH131 beta-Glucanase Catalytic Domain from Podospora anserina in Complex with Cellotriose
To be Published
|
|
4FEO
 
 | | Crystal structure of the AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | | Descriptor: | 9H-PURINE-2,6-DIAMINE, COBALT HEXAMMINE(III), U25A/A46G/C74U mutant of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain | | Authors: | Stoddard, C.D, Trausch, J.J, Widmann, J, Marcano, J, Knight, R, Batey, R.T. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleotides Adjacent to the Ligand-Binding Pocket are Linked to Activity Tuning in the Purine Riboswitch.
J.Mol.Biol., 425, 2013
|
|
6QPV
 
 | | Crystal structure of as isolated Y323A mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
