1YTJ
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | 分子名称: | PEPTIDE PRODUCT, SIV PROTEASE | | 著者 | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | 登録日 | 1996-08-01 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YTH
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | 分子名称: | HIV PROTEASE, PEPTIDE PRODUCT | | 著者 | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | 登録日 | 1996-08-01 | | 公開日 | 1997-03-12 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
3R43
 
 | | AKR1C3 complexed with mefenamic acid | | 分子名称: | 1,2-ETHANEDIOL, 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, Aldo-keto reductase family 1 member C3, ... | | 著者 | Squire, C.J, Teague, R.M, Yosaatmadja, L. | | 登録日 | 2011-03-17 | | 公開日 | 2012-03-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3OYP
 
 | | HCV NS3/4A in complex with ligand 3 | | 分子名称: | Non-structural protein 4A, Peptidomimetic inhibitor, Serine protease NS3, ... | | 著者 | Hagel, M, Niu, D, St.Martin, T, Sheets, M.P, Qiao, L, Bernard, H, Karp, R.M, Zhu, Z, Labenski, M.T, Chaturvedi, P.C, Nacht, M, Westlin, W.F, Petter, R.C, Singh, J. | | 登録日 | 2010-09-23 | | 公開日 | 2010-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Selective irreversible inhibition of a protease by targeting a noncatalytic cysteine.
Nat.Chem.Biol., 7, 2011
|
|
1YYQ
 
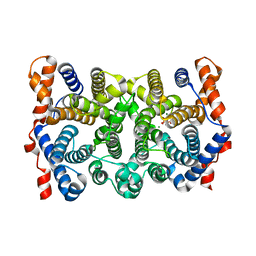 | | Y305F Trichodiene Synthase complexed with pyrophosphate | | 分子名称: | MAGNESIUM ION, PYROPHOSPHATE 2-, Trichodiene synthase | | 著者 | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | 登録日 | 2005-02-25 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1ZMT
 
 | | Structure of haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (R)-para-nitro styrene oxide, with a water molecule in the halide-binding site | | 分子名称: | (R)-PARA-NITROSTYRENE OXIDE, Haloalcohol dehalogenase HheC | | 著者 | de Jong, R.M, Tiesinga, J.J.W, Villa, A, Tang, L, Janssen, D.B, Dijkstra, B.W. | | 登録日 | 2005-05-10 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
3S12
 
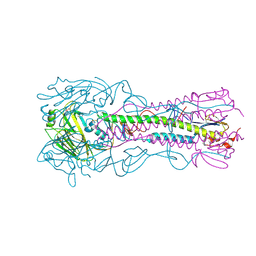 | | Crystal structure of H5N1 influenza virus hemagglutinin, strain YU562 crystal form 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | 著者 | DuBois, R.M, Zaraket, H, Reddivari, M, Heath, R.J, White, S.W, Russell, C.J. | | 登録日 | 2011-05-14 | | 公開日 | 2011-12-14 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity.
Plos Pathog., 7, 2011
|
|
3LZU
 
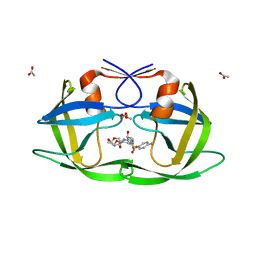 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | 著者 | Schiffer, C.A, Bandaranayake, R.M. | | 登録日 | 2010-03-01 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
1ZBX
 
 | | Crystal structure of a Orc1p-Sir1p complex | | 分子名称: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | 著者 | Hsu, H.C, Stillman, B, Xu, R.M. | | 登録日 | 2005-04-09 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1YYS
 
 | | Y305F Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4S)-7-azabisabolene | | 分子名称: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | 著者 | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | 登録日 | 2005-02-25 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | 分子名称: | halohydrin dehalogenase | | 著者 | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | 登録日 | 2005-05-10 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
3TZ0
 
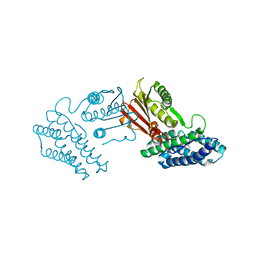 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/S-alpha-chloroisocaproate complex | | 分子名称: | (2S)-2-chloro-4-methylpentanoic acid, [3-methyl-2-oxobutanoate dehydrogenase [lipoamide]] kinase, mitochondrial | | 著者 | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | 登録日 | 2011-09-26 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1Z1Q
 
 | |
5YNL
 
 | | Crystal structure of a cold-adapted arginase from psychrophilic yeast, Glaciozyma antarctica | | 分子名称: | Arginase | | 著者 | Yusof, N.Y, Quay, D.H.X, Illias, R.M, Firdaus-Raih, M, Abu-Bakar, F.D, Mahadi, N.M, Murad, A.M.A. | | 登録日 | 2017-10-24 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | Crystal structure of a cold-adapted arginase from psychrophilic yeast, Glaciozyma antarctica
To Be Published
|
|
3UA3
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | 分子名称: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | 登録日 | 2011-10-20 | | 公開日 | 2011-12-14 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5YJ6
 
 | | The exoglucanase CelS from Clostridium thermocellum | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, Dockerin type I repeat-containing protein | | 著者 | Liu, Y.J, Liu, S.Y, Dong, S, Li, R.M, Feng, Y.G, Cui, Q. | | 登録日 | 2017-10-09 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum
Biotechnol Biofuels, 11, 2018
|
|
3OXV
 
 | | Crystal Structure of HIV-1 I50V, A71 Protease in Complex with the protease inhibitor amprenavir. | | 分子名称: | ACETATE ION, GLYCEROL, HIV-1 Protease, ... | | 著者 | Schiffer, C.A, Mittal, S, Bandaranayake, R.M. | | 登録日 | 2010-09-22 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3P28
 
 | |
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | 分子名称: | BACTERIOPHAGE PRD1 | | 著者 | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | 登録日 | 2001-04-13 | | 公開日 | 2001-12-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (25 Å) | | 主引用文献 | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | 登録日 | 2010-09-22 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
1C4F
 
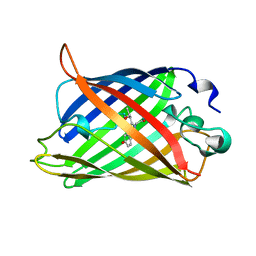 | | GREEN FLUORESCENT PROTEIN S65T AT PH 4.6 | | 分子名称: | GREEN FLUORESCENT PROTEIN | | 著者 | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | 登録日 | 1999-08-21 | | 公開日 | 1999-08-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
3OOI
 
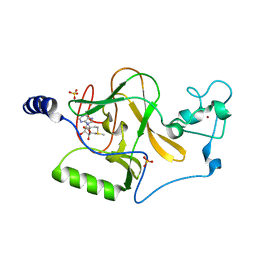 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | 著者 | Qiao, Q, Wang, M, Xu, R.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
1ZO8
 
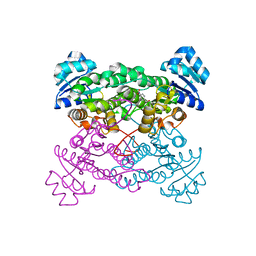 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | 分子名称: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | 著者 | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | 登録日 | 2005-05-12 | | 公開日 | 2005-10-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1YYT
 
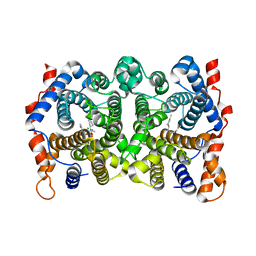 | | D100E Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4R)-7-azabisabolene | | 分子名称: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | 著者 | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | 登録日 | 2005-02-25 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
3RVK
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Q | | 分子名称: | Chemotaxis protein CheY, MANGANESE (II) ION | | 著者 | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | 登録日 | 2011-05-06 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
