5GWB
 
 | |
1DWS
 
 | | PHOTOLYZED CARBONMONOXY MYOGLOBIN (HORSE HEART) | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
1DWR
 
 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
6XYR
 
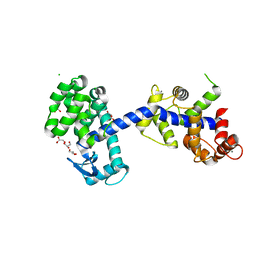 | | Structure of the T4Lnano fusion protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
1DTW
 
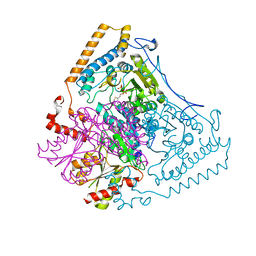 | | HUMAN BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE | | Descriptor: | BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE ALPHA SUBUNIT, BRANCHED-CHAIN ALPHA-KETO ACID DEHYDROGENASE BETA SUBUNIT, MAGNESIUM ION, ... | | Authors: | AEvarsson, A, Chuang, J.L, Wynn, R.M, Turley, S, Chuang, D.T, Hol, W.G.J. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human branched-chain alpha-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease.
Structure Fold.Des., 8, 2000
|
|
1E2B
 
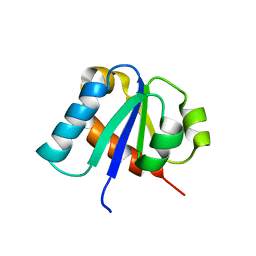 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
1DZ4
 
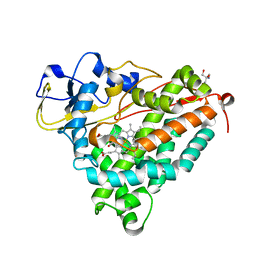 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ9
 
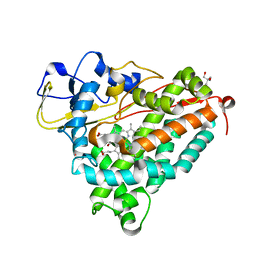 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
6QQ5
 
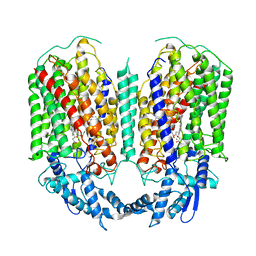 | | Cryo-EM structure of dimeric quinol dependent nitric oxide reductase (qNOR) from Alcaligenes xylosoxidans | | Descriptor: | CALCIUM ION, FE (III) ION, Nitric oxide reductase subunit B, ... | | Authors: | Gopalasingam, C.C, Johnson, R.M, Chiduza, G.N, Tosha, T, Yamamoto, M, Shiro, Y, Antonyuk, S.V, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Sci Adv, 5, 2019
|
|
6SXN
 
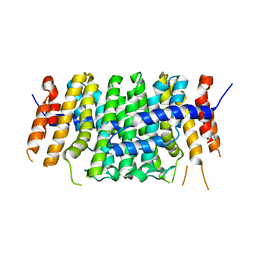 | | Crystal structure of P212121 apo form of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
1DWT
 
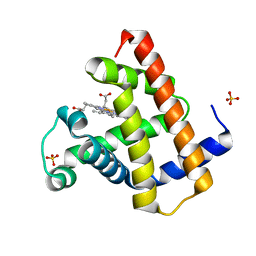 | | Photorelaxed horse heart MYOGLOBIN CO complex | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-12 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1DZC
 
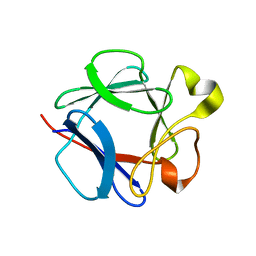 | | High resolution structure of acidic fibroblast growth factor. Mutant FGF-4-ALA-(24-154), 24 NMR structures | | Descriptor: | FIBROBLAST GROWTH FACTOR 1 | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
1DXC
 
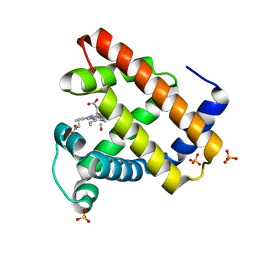 | | CO complex of Myoglobin Mb-YQR at 100K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Vallone, B, Cutruzzola, F, Travaglini-Allocatelli, C, Berendzen, J, Chu, K, Sweet, R.M, Schlichting, I. | | Deposit date: | 2000-01-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Role of Cavities in Protein Dynamics: Crystal Structure of a Novel Photolytic Intermediate of Myoglobin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5EE7
 
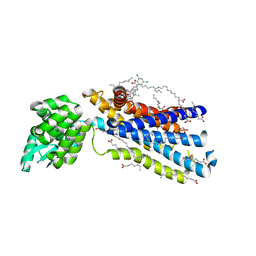 | | Crystal structure of the human glucagon receptor (GCGR) in complex with the antagonist MK-0893 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3-[[4-[(1~{S})-1-[3-[3,5-bis(chloranyl)phenyl]-5-(6-methoxynaphthalen-2-yl)pyrazol-1-yl]ethyl]phenyl]carbonylamino]propanoic acid, Glucagon receptor,Endolysin,Glucagon receptor, ... | | Authors: | Jazayeri, A, Dore, A.S, Lamb, D, Krishnamurthy, H, Southall, S.M, Baig, A.H, Bortolato, A, Koglin, M, Robertson, N.J, Errey, J.C, Andrews, S.P, Brown, A.J.H, Cooke, R.M, Weir, M, Marshall, F.H. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Extra-helical binding site of a glucagon receptor antagonist.
Nature, 533, 2016
|
|
1DNA
 
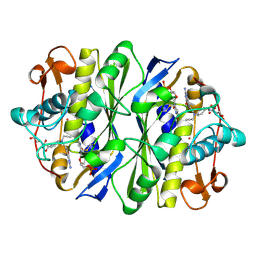 | | D221(169)N MUTANT DOES NOT PROMOTE OPENING OF THE COFACTOR IMIDAZOLIDINE RING | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Michelitsch, M.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D221 in thymidylate synthase controls conformation change, and thereby opening of the imidazolidine.
Biochemistry, 37, 1998
|
|
1DPO
 
 | | STRUCTURE OF RAT TRYPSIN | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stroud, R.M. | | Deposit date: | 1997-03-31 | | Release date: | 1997-07-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | 1.59 A structure of trypsin at 120 K: comparison of low temperature and room temperature structures.
Proteins, 10, 1991
|
|
7UK4
 
 | | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1DDU
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 AND 2',5'-DIDEOXYURIDINE (DDURD) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-5'DIDEOXYURIDINE, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
5EKF
 
 | |
6X3T
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
7TU6
 
 | | Structure of the L. blandensis dGTPase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU7
 
 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1DXD
 
 | | Photolyzed CO complex of Myoglobin Mb-YQR at 20K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Vallone, B, Cutruzzola, F, Travaglini-Allocatelli, C, Berendzen, J, Chu, K, Sweet, R.M, Schlichting, I. | | Deposit date: | 2000-01-03 | | Release date: | 2000-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Role of Cavities in Protein Dynamics: Crystal Structure of a Photolytic Intermediate of a Mutant Myoglobin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
