5TC3
 
 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP and GDP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
6MMD
 
 | | Photoactive Yellow Protein with 3,5-dichlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-30 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
1YYS
 
 | | Y305F Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
6MQD
 
 | | Myotoxin II from Bothrops moojeni complexed with Rosmarinic Acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, Basic phospholipase A2 homolog 2 | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Search for efficient inhibitors of myotoxic activity induced by ophidian phospholipase A2-like proteins using functional, structural and bioinformatics approaches.
Sci Rep, 9, 2019
|
|
1Z1Q
 
 | |
1ZBX
 
 | | Crystal structure of a Orc1p-Sir1p complex | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hsu, H.C, Stillman, B, Xu, R.M. | | Deposit date: | 2005-04-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1ZMO
 
 | | Apo structure of haloalcohol dehalogenase HheA of Arthrobacter sp. AD2 | | Descriptor: | halohydrin dehalogenase | | Authors: | de Jong, R.M, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray structure of the haloalcohol dehalogenase HheA from Arthrobacter sp. strain AD2: insight into enantioselectivity and halide binding in the haloalcohol dehalogenase family.
J.Bacteriol., 188, 2006
|
|
8BBX
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 in space group C222(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoprotease endo-Pro, TETRAETHYLENE GLYCOL, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8BV8
 
 | | Crystal structure of the phage Mu protein Mom inactive mutant S124A | | Descriptor: | Methylcarbamoylase mom | | Authors: | Silva, R.M.B, Slyvka, A, Lee, Y.J, Guan, C, Lund, S.R, Raleigh, E.A, Skowronek, K, Bochtler, M, Weigele, P.R. | | Deposit date: | 2022-12-08 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of the phage Mu protein Mom catalytic mutant S124A
To Be Published
|
|
8B57
 
 | | Structure of prolyl endoprotease from Aspergillus niger CBS 109712 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pijning, T, Vujicic-Zagar, A, Van der Laan, J.M, De Jong, R.M, Dijkstra, B.W. | | Deposit date: | 2022-09-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and time-resolved mechanistic investigations of protein hydrolysis by the acidic proline-specific endoprotease from Aspergillus niger.
Protein Sci., 33, 2024
|
|
8BWV
 
 | |
2AQ9
 
 | | Structure of E. coli LpxA with a bound peptide that is competitive with acyl-ACP | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Williams, A.H, Immormino, R.M, Gewirth, D.T, Raetz, C.R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7N1W
 
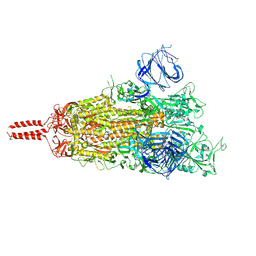 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1Y
 
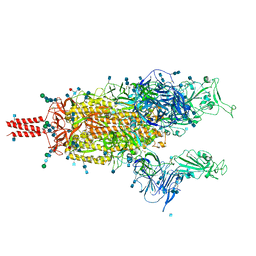 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1T
 
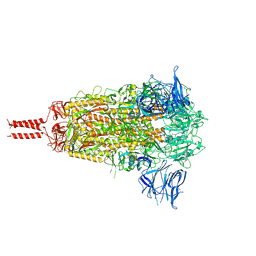 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1U
 
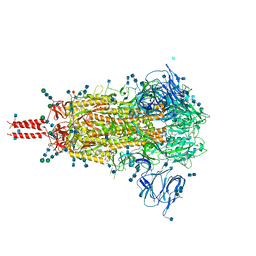 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
2B5Q
 
 | |
7N1Q
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1V
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
7N1X
 
 | | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Rawson, S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Chen, B. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants.
Science, 373, 2021
|
|
1VCA
 
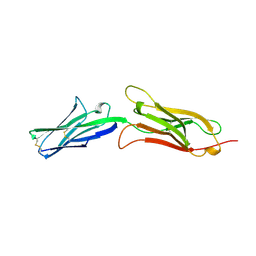 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
5V7C
 
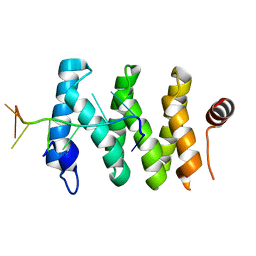 | |
5UYJ
 
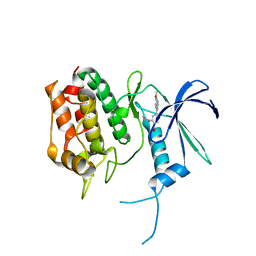 | | Crystal Structure of the Human CAMKK2B | | Descriptor: | 2-cyclopentyl-4-(7-methoxyquinolin-4-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, Drewry, D, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Human CAMKK2B
To Be Published
|
|
6M9U
 
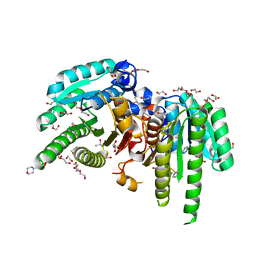 | | Structure of the apo-form of 20beta-Hydroxysteroid Dehydrogenase from Bifidobacterium adolescentis strain L2-32 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Mythen, S.M, Pollet, R.M, Koropatkin, N.M, Ridlon, J.M. | | Deposit date: | 2018-08-24 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of 20 beta-hydroxysteroid dehydrogenase fromBifidobacterium adolescentisstrain L2-32.
J.Biol.Chem., 294, 2019
|
|
5V4R
 
 | |
