2V6V
 
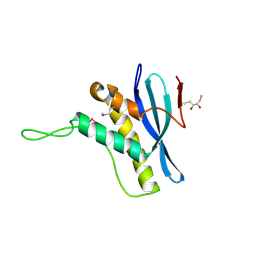 | | The structure of the Bem1p PX domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BUD EMERGENCE PROTEIN 1 | | Authors: | Stahelin, R.V, Karathanassis, D, Murray, D, Williams, R.L, Cho, W. | | Deposit date: | 2007-07-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Membrane Binding Analysis of the Phox Homology Domain of Bem1P: Basis of Phosphatidylinositol 4-Phosphate Specificity.
J.Biol.Chem., 282, 2007
|
|
3TIR
 
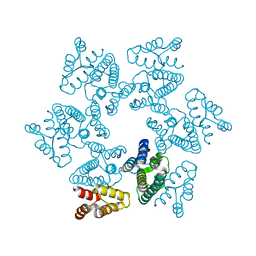 | |
2V14
 
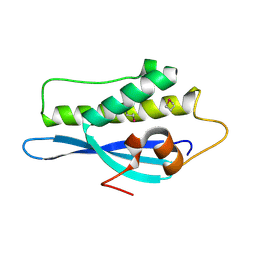 | | Kinesin 16B Phox-homology domain (KIF16B) | | Descriptor: | KINESIN-LIKE MOTOR PROTEIN C20ORF23 | | Authors: | Wilson, M.I, Williams, R.L, Cho, W, Hong, W, Blatner, N.R. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Novel Endosome Anchoring Activity of Kif16B Kinesin.
Embo J., 26, 2007
|
|
2V4E
 
 | | A non-cytotoxic DsRed variant for whole-cell labeling | | Descriptor: | RED FLUORESCENT PROTEIN DRFP583 | | Authors: | Strack, R.L, Strongin, D.E, Bhattacharyya, D, Tao, W, Berman, A, Broxmeyer, H.E, Keenan, R.J, Glick, B.S. | | Deposit date: | 2008-09-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Noncytotoxic Dsred Variant for Whole-Cell Labeling.
Nat.Methods, 5, 2008
|
|
3TVN
 
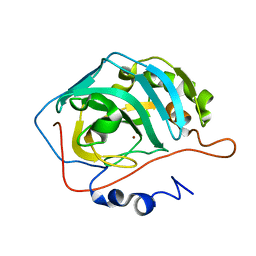 | | Human Carbonic Anhydrase II Proton Transfer Mutant | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Mikulski, R.L, West, D.M, Sippel, K.H, Avvaru, B.S, Chingkuang, T, McKenna, R. | | Deposit date: | 2011-09-20 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Water Networks in Fast Proton Transfer during Catalysis by Human Carbonic Anhydrase II.
Biochemistry, 52, 2013
|
|
3UM2
 
 | |
2VAD
 
 | | Monomeric red fluorescent protein, DsRed.M1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, RED FLUORESCENT PROTEIN, ... | | Authors: | Strongin, D.E, Bevis, B, Khuong, N, Downing, M.E, Strack, R.L, Sundaram, K, Glick, B.S, Keenan, R.J. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Rearrangements Near the Chromophore Influence the Maturation Speed and Brightness of Dsred Variants.
Protein Eng.Des.Sel., 20, 2007
|
|
3ULT
 
 | | Crystal structure of an ice-binding protein from the perennial ryegrass, Lolium perenne | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Ice recrystallization inhibition protein-like protein | | Authors: | Middleton, A.J, Faucher, F, Campbell, R.L, Davies, P.L. | | Deposit date: | 2011-11-11 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antifreeze protein from freeze-tolerant grass has a beta-roll fold with an irregularly structured ice-binding site.
J.Mol.Biol., 416, 2012
|
|
3ULY
 
 | |
2PET
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2. | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PF6
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2 | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2V4Q
 
 | | Post-insertion complex of the Y-family DNA polymerase Dpo4 with M1dG containing template DNA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*C M1GP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Eoff, R.L, Stafford, J.B, Szekely, J, Rizzo, C.J, Egli, M, Guengerich, F.P, Marnett, L.J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Analysis of Sulfolobus Solfataricus Y-Family DNA Polymerase Dpo4-Catalyzed Bypass of the Malondialdehyde-Deoxyguanosine Adduct.
Biochemistry, 48, 2009
|
|
2V4L
 
 | | complex of human phosphoinositide 3-kinase catalytic subunit gamma (p110 gamma) with PIK-284 | | Descriptor: | 3-[4-AMINO-1-(1-METHYLETHYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL]PHENOL, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Apsel, B, Gonzalez, B, Blair, J.A, Nazif, T.M, Feldman, M.E, Williams, R.L, Shokat, K.M, Knight, Z.A. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeted Polypharmacology: Discovery of Dual Inhibitors of Tyrosine and Phosphoinositide Kinases.
Nat.Chem.Biol., 4, 2008
|
|
3VBL
 
 | | Crystal Structure of the S84C mutant of AntD, an N-acyltransferase from Bacillus cereus in complex with dTDP-4-amino-4,6-dideoxyglucose and Coenzyme A | | Descriptor: | BICARBONATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kubiak, R.L, Holden, H.M. | | Deposit date: | 2012-01-02 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of AntD: An N-Acyltransferase Involved in the Biosynthesis of d-Anthrose.
Biochemistry, 51, 2012
|
|
1TQO
 
 | |
1U5A
 
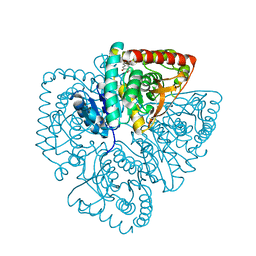 | | Plasmodium falciparum lactate dehydrogenase complexed with 3,5-dihydroxy-2-naphthoic acid | | Descriptor: | 3,7-DIHYDROXY-2-NAPHTHOIC ACID, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-07-27 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase.
Mol.Biochem.Parasitol., 142, 2005
|
|
1TZG
 
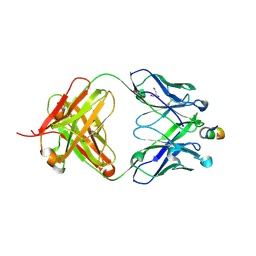 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
1TWZ
 
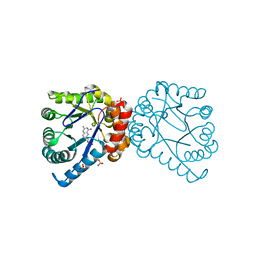 | | Dihydropteroate Synthetase, With Bound Substrate Analogue PtP, From Bacillus anthracis | | Descriptor: | DHPS, Dihydropteroate synthase, PTERIN-6-YL-METHYL-MONOPHOSPHATE, ... | | Authors: | Babaoglu, K, Qi, J, Lee, R.L, White, S.W. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of 7,8-Dihydropteroate Synthase from Bacillus anthracis; Mechanism and Novel Inhibitor Design.
STRUCTURE, 12, 2004
|
|
1UZX
 
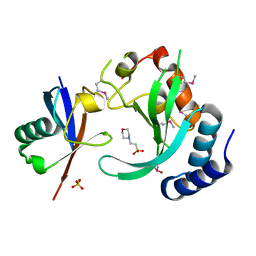 | | A complex of the Vps23 UEV with ubiquitin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UBIQUITIN, ... | | Authors: | Teo, H, Williams, R.L. | | Deposit date: | 2004-03-18 | | Release date: | 2004-03-30 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Endosomal Sorting Complex Required for Transport (Escrt-I) Recognition of Ubiquitinated Proteins
J.Biol.Chem., 279, 2004
|
|
1W2F
 
 | | Human Inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine | | Descriptor: | INOSITOL-TRISPHOSPHATE 3-KINASE A, SULFATE ION | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1WA1
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1W2C
 
 | | Human Inositol (1,4,5) trisphosphate 3-kinase complexed with Mn2+/AMPPNP/Ins(1,4,5)P3 | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, INOSITOL-TRISPHOSPHATE 3-KINASE A, MANGANESE (II) ION, ... | | Authors: | Gonzalez, B, Schell, M.J, Irvine, R.F, Williams, R.L. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Human Inositol 1,4,5-Trisphosphate 3-Kinase; Substrate Binding Reveals Why It is not a Phosphoinositide 3-Kinase
Mol.Cell, 15, 2004
|
|
1WKO
 
 | |
1WA0
 
 | | Crystal Structure Of W138H Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
1WA2
 
 | | Crystal Structure Of H313Q Mutant Of Alcaligenes Xylosoxidans Nitrite Reductase with nitrite bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE,, ... | | Authors: | Barrett, M.L, Harris, R.L, Antonyuk, S.V, Strange, R.W, Hough, M.A, Eady, R.R, Sawers, G, Hasnain, S.S. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights Into Redox Partner Interactions and Substrate Binding in Nitrite Reductase from Alcaligenes Xylosoxidans: Crystal Structures of the Trp138His and His313Gln Mutants
Biochemistry, 43, 2004
|
|
