1EAI
 
 | | COMPLEX OF ASCARIS CHYMOTRPSIN/ELASTASE INHIBITOR WITH PORCINE ELASTASE | | Descriptor: | PROTEIN (CHYMOTRYPSIN/ELASTASE ISOINHIBITOR 1), PROTEIN (ELASTASE) | | Authors: | Huang, K, Strynadka, N.C.J, Bernard, V.D, Peanasky, R.J, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the complex of Ascaris chymotrypsin/elastase inhibitor with porcine elastase.
Structure, 2, 1994
|
|
1EBL
 
 | | THE 1.8 A CRYSTAL STRUCTURE AND ACTIVE SITE ARCHITECTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE III (FABH) FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL-ACP SYNTHASE III, COENZYME A | | Authors: | Davies, C, Heath, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2000-01-24 | | Release date: | 2000-02-11 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A crystal structure and active-site architecture of beta-ketoacyl-acyl carrier protein synthase III (FabH) from escherichia coli.
Structure Fold.Des., 8, 2000
|
|
1HEU
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1HET
 
 | | atomic X-ray structure of liver alcohol dehydrogenase containing a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-25 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1H4Z
 
 | | Structure of the Anti-Sigma Factor Antagonist SpoIIAA in its Unphosphorylated Form | | Descriptor: | ANTI-SIGMA F FACTOR ANTAGONIST | | Authors: | Seavers, P.R, Lewis, R.J, Brannigan, J.A, Verschueren, K.H.G, Murshudov, G.N, Wilkinson, A.J. | | Deposit date: | 2001-05-16 | | Release date: | 2001-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the Bacillus Cell Fate Determinant Spoiiaa in Phosphorylated and Unphosphorylated Forms
Structure, 9, 2001
|
|
1HF3
 
 | | ATOMIC X-RAY STRUCTURE OF LIVER ALCOHOL DEHYDROGENASE CONTAINING Cadmium and a hydroxide adduct to NADH | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ALCOHOL DEHYDROGENASE E CHAIN, CADMIUM ION, ... | | Authors: | Meijers, R, Morris, R.J, Adolph, H.W, Merli, A, Lamzin, V.S, Cedergen-Zeppezauer, E.S. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | On the Enzymatic Activation of Nadh
J.Biol.Chem., 276, 2001
|
|
1F16
 
 | |
1H0W
 
 | | Human cyclin dependent protein kinase 2 in complex with the inhibitor 2-Amino-6-[cyclohex-3-enyl]methoxypurine | | Descriptor: | 1-AMINO-6-CYCLOHEX-3-ENYLMETHYLOXYPURINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Gibson, A.E, Arris, C.E, Bentley, J, Boyle, F.T, Curtin, N.J, Davies, T.G, Endicott, J.A, Golding, B.T, Grant, S, Griffin, R.J, Jewsbury, P, Johnson, L.N, Mesguiche, V, Newell, D.R, Noble, M.E.M, Tucker, J.A, Whitfield, H.J. | | Deposit date: | 2002-06-27 | | Release date: | 2003-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the ATP Ribose-Binding Domain of Cyclin-Dependent Kinases 1 and 2 with O(6)-Substituted Guanine Derivatives
J.Med.Chem., 45, 2002
|
|
1HMU
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1U0H
 
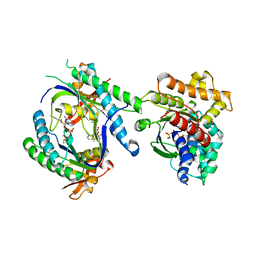 | | STRUCTURAL BASIS FOR THE INHIBITION OF MAMMALIAN ADENYLYL CYCLASE BY MANT-GTP | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | Authors: | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
3NT5
 
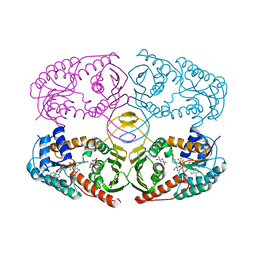 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9006 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTO
 
 | |
3OA9
 
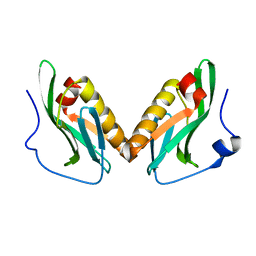 | | Effector domain of influenza A/Duck/Albany/76 NS1 | | Descriptor: | Non-structural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3NT2
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3003 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
4D8S
 
 | | Influenza NA in complex with antiviral compound | | Descriptor: | CALCIUM ION, Neuraminidase, pentan-3-yl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid | | Authors: | Kerry, P.S, Russell, R.J.M.R. | | Deposit date: | 2012-01-11 | | Release date: | 2013-02-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Exploring the interactions of unsaturated glucuronides with influenza virus sialidase.
J.Med.Chem., 55, 2012
|
|
2VL0
 
 | |
3NT4
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
3NTQ
 
 | |
3O9S
 
 | | Effector domain of influenza A/PR/8/34 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9R
 
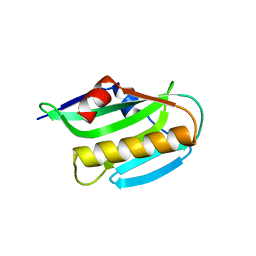 | | Effector domain of NS1 from influenza A/PR/8/34 containing a W187A mutation | | Descriptor: | Nonstructural protein 1 | | Authors: | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | Deposit date: | 2010-08-04 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
2VW2
 
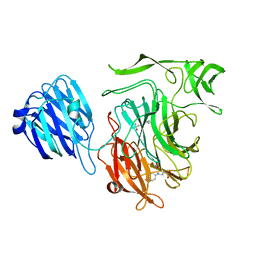 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VW0
 
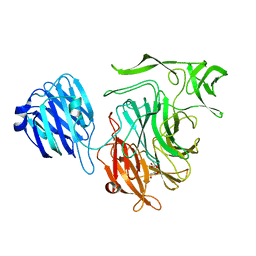 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
2VC6
 
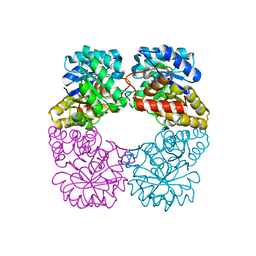 | | Structure of MosA from S. meliloti with pyruvate bound | | Descriptor: | DIHYDRODIPICOLINATE SYNTHASE | | Authors: | Phenix, C.P, Nienaber, K.H, Tam, P.H, Delbaere, L.T.J, Palmer, D.R.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, functional and calorimetric investigation of MosA, a dihydrodipicolinate synthase from Sinorhizobium meliloti l5-30, does not support involvement in rhizopine biosynthesis.
Chembiochem, 9, 2008
|
|
4E80
 
 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | Poly(A) RNA polymerase protein cid1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4E8F
 
 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | ACETATE ION, GLYCEROL, Poly(A) RNA polymerase protein cid1 | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
