3PZ6
 
 | | The crystal structure of GlLeuRS-CP1 | | Descriptor: | Leucyl-tRNA synthetase | | Authors: | Liu, R.J, Du, D.H, Wang, E.D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Peripheral insertion modulates the editing activity of the isolated CP1 domain of leucyl-tRNA synthetase
Biochem.J., 440, 2011
|
|
1XQM
 
 | | Variations on the GFP chromophore scaffold: A fragmented 5-membered heterocycle revealed in the 2.1A crystal structure of a non-fluorescent chromoprotein | | Descriptor: | ACETIC ACID, kindling fluorescent protein | | Authors: | Wilmann, P.G, Petersen, J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variations on the GFP chromophore: A polypeptide fragmentation within the chromophore revealed in the 2.1-A crystal structure of a nonfluorescent chromoprotein from Anemonia sulcata
J.Biol.Chem., 280, 2005
|
|
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
5CMG
 
 | | GTA mutant with mercury- E303C | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M, Blackler, R.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
4P66
 
 | | Electrostatics of Active Site Microenvironments of E. coli DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, METHOTREXATE, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
5UFB
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) Tn4-22 (Apo) | | Descriptor: | Tn4-22 | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Cummings, R.D, Cooper, M.D, Herrin, B.R, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
5UFF
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 with Fucose(alpha-1-2)Lactose bound | | Descriptor: | RBC36, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
6BDY
 
 | |
5CMJ
 
 | | GTA mutant with mercury - E303Q | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
3PZV
 
 | | C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
1YKS
 
 | | Crystal structure of yellow fever virus NS3 helicase | | Descriptor: | Genome polyprotein [contains: Flavivirin protease NS3 catalytic subunit] | | Authors: | Wu, J, Bera, A.K, Kuhn, R.J, Smith, J.L. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing.
J.Virol., 79, 2005
|
|
5CMH
 
 | | GTA mutant with mercury - E303D | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
1YMT
 
 | | Mouse SF-1 LBD | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, Nuclear receptor 0B2, Steroidogenic factor 1 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, Juzumiene, D, Bynum, J.M, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
1YS2
 
 | | Burkholderia cepacia lipase complexed with hexylphosphonic acid (S) 2-methyl-3-phenylpropyl ester | | Descriptor: | CALCIUM ION, HEXYLPHOSPHONIC ACID (S)-2-METHYL-3-PHENYLPROPYL ESTER, Lipase | | Authors: | Mezzetti, A, Schrag, J.D, Cheong, C.S, Kazlauskas, R.J. | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mirror-Image Packing in Enantiomer Discrimination Molecular Basis for the Enantioselectivity of B.cepacia Lipase toward 2-Methyl-3-Phenyl-1-Propanol.
Chem.Biol., 12, 2005
|
|
6BAV
 
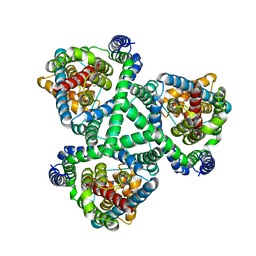 | | Crystal Structure of GltPh R397C in complex with S-Benzyl-L-Cysteine | | Descriptor: | BENZYLCYSTEINE, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
5CQO
 
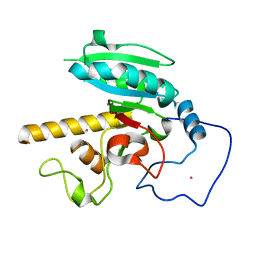 | | GTB mutant with mercury - E303D | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-21 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
3RKG
 
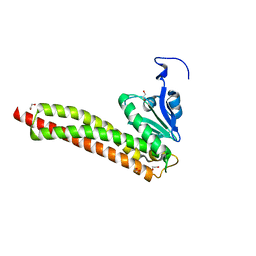 | | Structural and Functional Characterization of the Yeast Mg2+ Channel Mrs2 | | Descriptor: | 1,2-ETHANEDIOL, Magnesium transporter MRS2, mitochondrial | | Authors: | Khan, M.B, Sponder, G, Sjoeblom, B, Svidova, S, Schweyen, R.J, Carugo, O, Djinovic-Carugo, K. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural and functional characterization of the N-terminal domain of the yeast Mg(2+) channel Mrs2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4P68
 
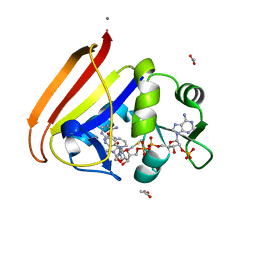 | | Electrostatics of Active Site Microenvironments for E. coli DHFR | | Descriptor: | ACETATE ION, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Liu, C.T, Layfield, J.P, Stewart III, R.J, French, J.B, Hanoian, P, Asbury, J.B, Hammes-Schiffer, S, Benkovic, S.J. | | Deposit date: | 2014-03-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the electrostatics of active site microenvironments along the catalytic cycle for Escherichia coli dihydrofolate reductase.
J.Am.Chem.Soc., 136, 2014
|
|
5CXU
 
 | |
1YTZ
 
 | | Crystal structure of skeletal muscle troponin in the Ca2+-activated state | | Descriptor: | ALPHA-[4-(1,1,3,3 - TETRAMETHYLBUTYL)PHENYL]-OMEGA-HYDROXY-POLY(OXY-1,2-ETHANEDIYL), CALCIUM ION, Troponin C, ... | | Authors: | Vinogradova, M.V, Stone, D.B, Malanina, G.G, Karatzaferi, C, Cooke, R, Mendelson, R.A, Fletterick, R.J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca2+-regulated structural changes in troponin
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6CFB
 
 | | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti | | Descriptor: | barrettide A | | Authors: | Rosengren, K.J, Carstens, B.B, Clark, R.J, Goransson, U. | | Deposit date: | 2018-02-14 | | Release date: | 2018-03-21 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti.
J. Nat. Prod., 78, 2015
|
|
2BOL
 
 | | CRYSTAL STRUCTURE AND ASSEMBLY OF TSP36, A METAZOAN SMALL HEAT SHOCK PROTEIN | | Descriptor: | SMALL HEAT SHOCK PROTEIN, SULFATE ION | | Authors: | Stamler, R.J, Kappe, G, Boelens, W.C, Slingsby, C. | | Deposit date: | 2005-04-12 | | Release date: | 2005-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Wrapping the Alpha-Crystallin Domain Fold in a Chaperone Assembly.
J.Mol.Biol., 353, 2005
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
1YS1
 
 | | Burkholderia cepacia lipase complexed with hexylphosphonic acid (R)-2-methyl-3-phenylpropyl ester | | Descriptor: | CALCIUM ION, HEXYLPHOSPHONIC ACID (R)-2-METHYL-3-PHENYLPROPYL ESTER, Lipase | | Authors: | Mezzetti, A, Schrag, J.D, Cheong, C.S, Kazlauskas, R.J. | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing in Enantiomer Discrimination Molecular Basis for the Enantioselectivity of B.cepacia Lipase toward 2-Methyl-3-Phenyl-1-Propanol.
Chem.Biol., 12, 2005
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
