8RXT
 
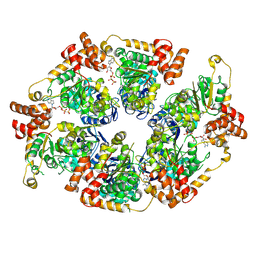 | | ComM helicase hexamer in abscence o DNA | | Descriptor: | Competence related protein ComM, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Talachia Rosa, L, Fronzes, R. | | Deposit date: | 2024-02-07 | | Release date: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural insights into the mechanism of DNA branch migration during homologous recombination in bacteria.
Embo J., 2024
|
|
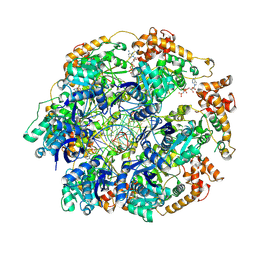
8RXC
 
 | |
8RXD
 
 | |
4BN6
 
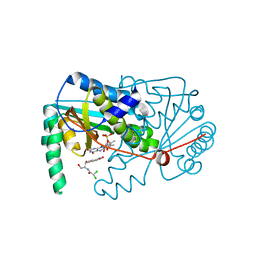 | |
2H8H
 
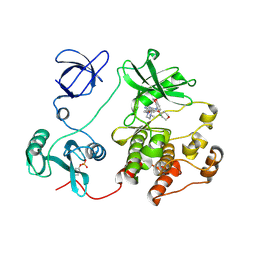 | | Src kinase in complex with a quinazoline inhibitor | | Descriptor: | N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Otterbein, L.R, Norman, R, Pauptit, R.A, Rowsell, S, Breed, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor.
J.Med.Chem., 49, 2006
|
|
3VF1
 
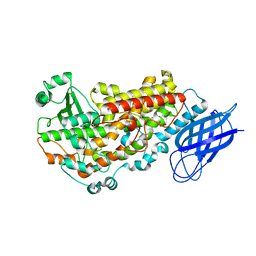 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
1J8Q
 
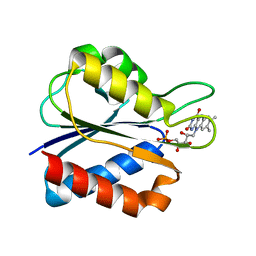 | | Low Temperature (100K) Crystal Structure of Flavodoxin D. vulgaris Wild-type at 1.35 Angstrom Resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Bombieri, G, Meneghetti, F, Gilardi, G, Sadeghi, S.J, Cavazzini, D, Rossi, G.L. | | Deposit date: | 2001-05-22 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Comparison of the refined crystal structures of wild-type (1.34 A) flavodoxin from Desulfovibrio vulgaris and the S35C mutant (1.44 A) at 100 K.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4IPF
 
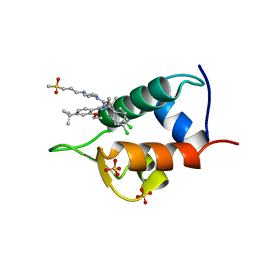 | | The 1.7A crystal structure of humanized Xenopus MDM2 with RO5045337 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Graves, B.J, Lukacs, C, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models.
Cancer Res., 73, 2013
|
|
3VB8
 
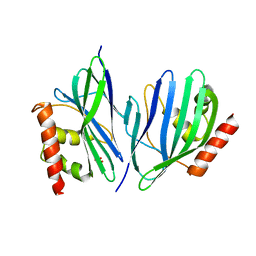 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR43 | | Descriptor: | Engineered protein, SULFATE ION | | Authors: | Seetharaman, J, Su, M, Procko, E, Baker, D, Ciccosanti, C, Sahdev, S, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-31 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Computational design of a protein-based enzyme inhibitor.
J.Mol.Biol., 425, 2013
|
|
8ZDZ
 
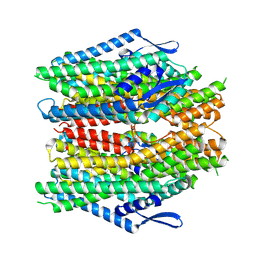 | |
2YEP
 
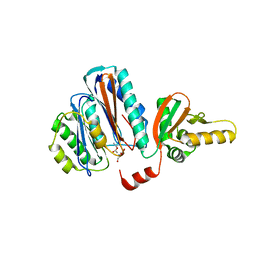 | | STRUCTURE OF AN N-TERMINAL NUCLEOPHILE (NTN) HYDROLASE, OAT2, IN COMPLEX WITH GLUTAMATE | | Descriptor: | ACETATE ION, GLUTAMATE N-ACETYLTRANSFERASE 2 ALPHA CHAIN, GLUTAMATE N-ACETYLTRANSFERASE 2 BETA CHAIN, ... | | Authors: | Chowdhury, R, Iqbal, A, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-29 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Analyses Reveal How Ornithine Acetyl Transferase Binds Acidic and Basic Amino Acid Substrates.
Org.Biomol.Chem., 9, 2011
|
|
8ZE3
 
 | |
2YOE
 
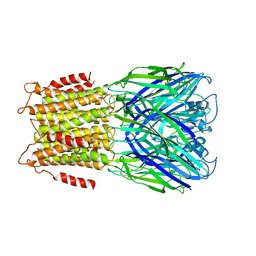 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with GABA and flurazepam | | Descriptor: | CYS-LOOP LIGAND-GATED ION CHANNEL, Flurazepam, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Spurny, R, Brams, M, Nury, H, Legrand, P, Ulens, C. | | Deposit date: | 2012-10-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Pentameric Ligand-Gated Ion Channel Elic is Activated by Gaba and Modulated by Benzodiazepines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1J7Z
 
 | | Osmolyte Stabilization of Ribonuclease | | Descriptor: | RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 2001-05-19 | | Release date: | 2001-06-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
1J81
 
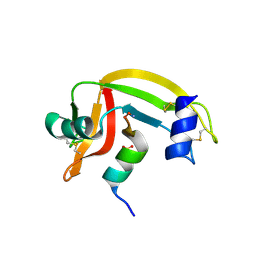 | | Osmolyte Stabilization of RNase | | Descriptor: | RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 2001-05-19 | | Release date: | 2001-06-06 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
4BZK
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
2GTO
 
 | |
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
4IVR
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with IN52/10 | | Descriptor: | 2-[(2,6-dimethoxy-5-methylpyrimidin-4-yl)methylidene]propane-1,3-diol, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
2Y66
 
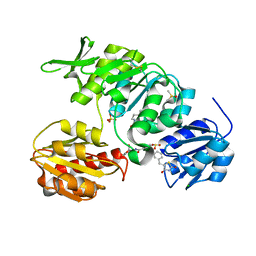 | | New 5-Benzylidenethiazolidine-4-one Inhibitors of Bacterial MurD Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation | | Descriptor: | (2R)-2-[[3-[[3-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]phenoxy]methyl]phenyl]carbonylamino]pentanedioic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Zidar, N, Tomasic, T, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | New 5-Benzylidenethiazolidin-4-One Inhibitors of Bacterial Murd Ligase: Design, Synthesis, Crystal Structures, and Biological Evaluation.
Eur.J.Med.Chem, 46, 2011
|
|
5QHK
 
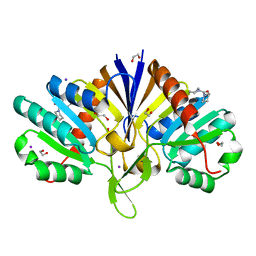 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000010a | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QI7
 
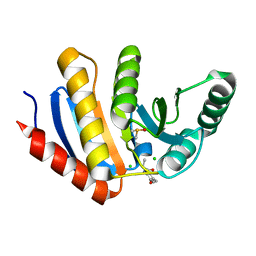 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000506a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1IJZ
 
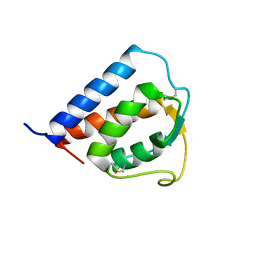 | |
4BOP
 
 | | Structure of OTUD1 OTU domain | | Descriptor: | OTU DOMAIN-CONTAINING PROTEIN 1, PHOSPHATE ION | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
5QIY
 
 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102954 | | Descriptor: | N'-acetyl-2-chlorobenzohydrazide, UNKNOWN LIGAND, Ubiquitin thioesterase OTUB2 | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
