5EYF
 
 | | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, Glutamate ABC superfamily ATP binding cassette transporter, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Solute-binding Protein from Enterococcus faecium with Bound Glutamate
To Be Published
|
|
5F6Y
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3O1Y
 
 | | Electron transfer complexes: Experimental mapping of the redox-dependent cytochrome c electrostatic surface | | Descriptor: | Cytochrome c, HEME C, NITRATE ION | | Authors: | De March, M, De Zorzi, R, Demitri, N, Gabbiani, C, Guerri, A, Casini, A, Messori, L, Geremia, S. | | Deposit date: | 2010-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nitrate as a probe of cytochrome c surface: crystallographic identification of crucial "hot spots" for protein-protein recognition.
J. Inorg. Biochem., 135, 2014
|
|
5EZ7
 
 | | Crystal structure of the FAD dependent oxidoreductase PA4991 from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, flavoenzyme PA4991 | | Authors: | Jacewicz, A, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the flavoenzyme PA4991 from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EZK
 
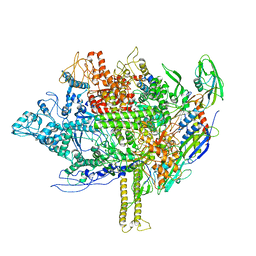 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
6SNK
 
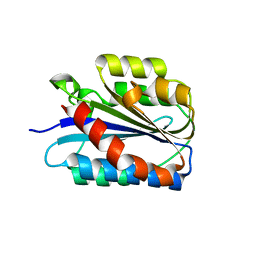 | | Crystal structure of the Collagen VI alpha3 N2 domain | | Descriptor: | Collagen alpha-3(VI) chain | | Authors: | Gebauer, J.M, Degefa, H.S, Paulsson, M, Wagener, R, Baumann, U. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a collagen VI alpha 3 chain VWA domain array: adaptability and functional implications of myopathy causing mutations.
J.Biol.Chem., 295, 2020
|
|
5F1Y
 
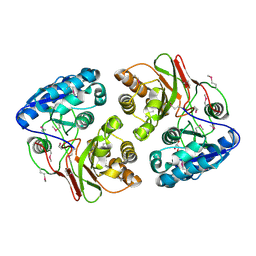 | |
6SP1
 
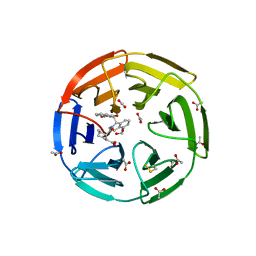 | | KEAP1 IN COMPLEX WITH COMPOUND 6 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclohexane-1-carboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
6SV9
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x40) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
2ZLV
 
 | | Horse methemoglobin high salt, pH 7.0 (79% relative humidity) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kaushal, P.S, Sankaranarayanan, R, Vijayan, M. | | Deposit date: | 2008-04-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Water-mediated variability in the structure of relaxed-state haemoglobin
Acta Crystallogr.,Sect.F, 64, 2008
|
|
6SE1
 
 | | Structure of Salmonella ser. Paratyphi A lipopolysaccharide acetyltransferase periplasmic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative lipopolysaccharide modification acyltransferase, SULFATE ION | | Authors: | Tindall, S.N, Pearson, C, Herman, R, Jenkins, H.T, Thomas, G.H, Potts, J.R, Van der Woude, M. | | Deposit date: | 2019-07-29 | | Release date: | 2020-08-26 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Acetylation of Surface Carbohydrates in Bacterial Pathogens Requires Coordinated Action of a Two-Domain Membrane-Bound Acyltransferase.
Mbio, 11, 2020
|
|
6SGE
 
 | | Crystal structure of Human RHOB-GTP in complex with nanobody B6 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nanobody B6, ... | | Authors: | Soulie, S, Gence, R, Cabantous, S, Lajoie-Mazenc, I, Favre, G, Pedelacq, J.D. | | Deposit date: | 2019-08-04 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Targeted Protein Degradation Cell-Based Screening for Nanobodies Selective toward the Cellular RHOB GTP-Bound Conformation.
Cell Chem Biol, 26, 2019
|
|
6SH4
 
 | |
6SJL
 
 | |
6SC4
 
 | | Gamma-Carbonic Anhydrase from the Haloarchaeon Halobacterium sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Vogler, M, Karan, R, Renn, D, Vancea, A, Vielberg, V.-T, Groetzinger, S.W, DasSarma, P, Das Sarma, S, Eppinger, J, Groll, M, Rueping, M. | | Deposit date: | 2019-07-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Active Site Engineering of a Halophilic gamma-Carbonic Anhydrase.
Front Microbiol, 11, 2020
|
|
6SKI
 
 | |
5FAG
 
 | |
5FBW
 
 | | PI4KB in complex with Rab11 and the MI369 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | PI4KB in complex with Rab11 and the MI369 Inhibitor
To Be Published
|
|
5FD5
 
 | | manganese uptake regulator | | Descriptor: | 1,2-ETHANEDIOL, Ferric uptake regulation protein, SULFATE ION | | Authors: | Bellini, D, Lebedev, A, Keegan, R, Walsh, M.A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of an apo metal-free manganese uptake regulator, mur
To Be Published
|
|
5FY7
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, MANGANESE (II) ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Succinate
To be Published
|
|
6SSP
 
 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a NAM nanobody | | Descriptor: | CALCIUM ION, Cys-loop ligand-gated ion channel, NANOBODY 21, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
5FYS
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with D-2-hydroxyglutarate | | Descriptor: | (2R)-2-hydroxypentanedioic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with D-2-Hydroxyglutarate
To be Published
|
|
6SV6
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x38) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-17 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
5FZI
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC3095 | | Descriptor: | 1,2-ETHANEDIOL, 6-oxo-2-[(2-oxo-2-phenylethyl)sulfanyl]-1,6-dihydropyrimidine-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Rotili, D, Mai, A, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc3095
To be Published
|
|
6SVN
 
 | | Reference structure of bovine trypsin (even frames of crystal x28) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
