7Q37
 
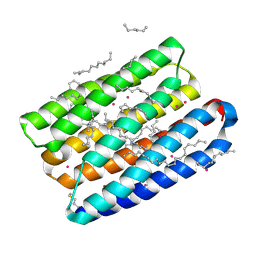 | | Crystal structure of proton pump MAR rhodopsin pressurized with krypton | | Descriptor: | Bacteriorhodopsin, EICOSANE, HEXANE, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
4XFE
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, SULFATE ION, TRAP dicarboxylate transporter subunit DctP, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-26 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a TRAP periplasmic solute binding protein from Pseudomonas putida F1 (Pput_1203), Target EFI-500184, with bound D-glucuronate
To be published
|
|
7Q38
 
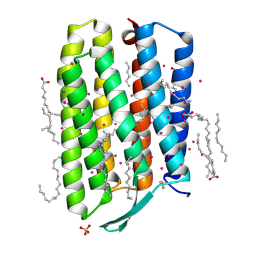 | | Crystal structure of the mutant bacteriorhodopsin pressurized with argon | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ARGON, Bacteriorhodopsin, ... | | Authors: | Melnikov, I, Rulev, M, Astashkin, R, Kovalev, K, Carpentier, P, Gordeliy, V, Popov, A. | | Deposit date: | 2021-10-27 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-pressure crystallography shows noble gas intervention into protein-lipid interaction and suggests a model for anaesthetic action.
Commun Biol, 5, 2022
|
|
4Y1H
 
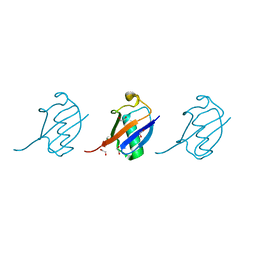 | | Crystal structure of K33 linked tri-Ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin-40S ribosomal protein S27a | | Authors: | Kristariyanto, Y.A, Abdul Rehman, S.A, Choi, S.Y, Ritorto, S, Campbell, D.G, Morrice, N.A, Toth, R, Kulathu, Y. | | Deposit date: | 2015-02-07 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Assembly and structure of Lys33-linked polyubiquitin reveals distinct conformations.
Biochem.J., 467, 2015
|
|
6ONF
 
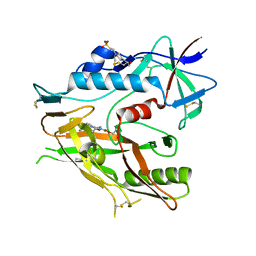 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A02. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6OWZ
 
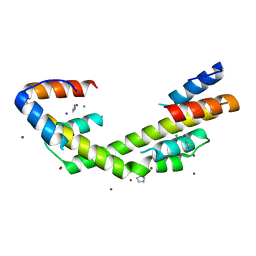 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7PGX
 
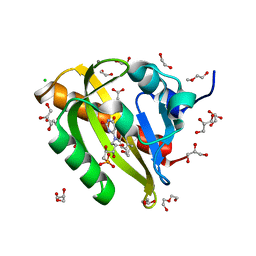 | | Structure of dark-adapted AsLOV2 wild type | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.001 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
8QWR
 
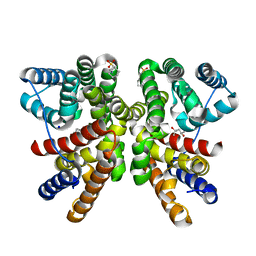 | | Crystal structure of CotB2 variant V80L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclooctat-9-en-7-ol synthase, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CotB2 variant V80L
To Be Published
|
|
8QLF
 
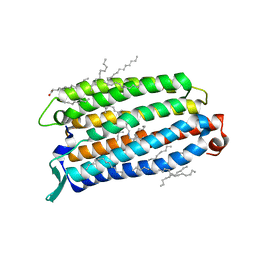 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
1F4Z
 
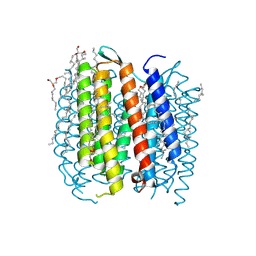 | | BACTERIORHODOPSIN-M PHOTOINTERMEDIATE STATE OF THE E204Q MUTANT AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Luecke, H, Schobert, B, Cartailler, J.P, Richter, H.T, Rosengarth, A, Needleman, R, Lanyi, J.K. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coupling photoisomerization of retinal to directional transport in bacteriorhodopsin.
J.Mol.Biol., 300, 2000
|
|
7PGZ
 
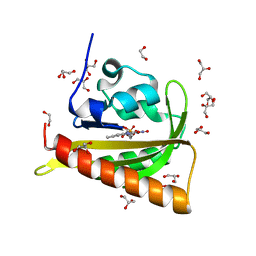 | | Structure of dark-adapted AsLOV2 Q513L | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
7EET
 
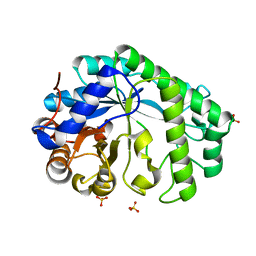 | | Mannanase KMAN from Klebsiella oxytoca KUB-CW2-3 | | Descriptor: | Mannanase KMAN, SULFATE ION | | Authors: | Pongsapipatana, N, Charoenwattanasatien, R, Pramanpol, N, Nitisinprasert, S, Keawsompong, S. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystallization, structural characterization and kinetic analysis of a GH26 beta-mannanase from Klebsiella oxytoca KUB-CW2-3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7PH0
 
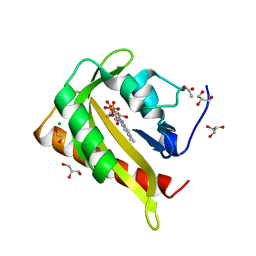 | | Structure of light-adapted AsLOV2 Q513L | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Gelfert, R, Weyand, M, Moeglich, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.979 Å) | | Cite: | Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine.
Nat Commun, 13, 2022
|
|
6OSU
 
 | | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase (Penicillin binding protein) family protein | | Authors: | Kim, Y, Stogios, P, Skarina, T, Di, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis
To Be Published
|
|
1F3E
 
 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
4XND
 
 | |
6OTI
 
 | |
6O8Y
 
 | |
6OTM
 
 | |
6OTP
 
 | |
4XOD
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 | | Descriptor: | FimG protein, FimH protein | | Authors: | Jakob, R.P, Sauer, M.M, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
6OUI
 
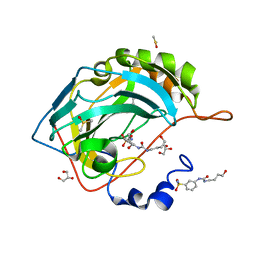 | |
1F4S
 
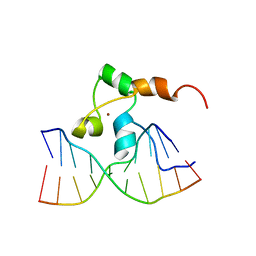 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-09 | | Release date: | 2001-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
1F5E
 
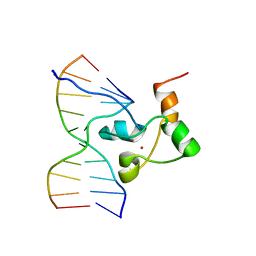 | | STRUCTURE OF TRANSCRIPTIONAL FACTOR ALCR IN COMPLEX WITH A TARGET DNA | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*CP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*GP*CP*AP*CP*G)-3'), ETHANOL REGULON TRANSCRIPTIONAL FACTOR, ... | | Authors: | Cahuzac, B, Cerdan, R, Felenbok, B, Guittet, E. | | Deposit date: | 2000-06-14 | | Release date: | 2001-09-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an AlcR-DNA complex sheds light onto the unique tight and monomeric DNA binding of a Zn(2)Cys(6) protein.
Structure, 9, 2001
|
|
6OXM
 
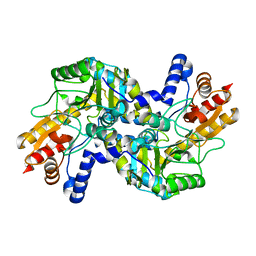 | |
