5MP1
 
 | |
5C56
 
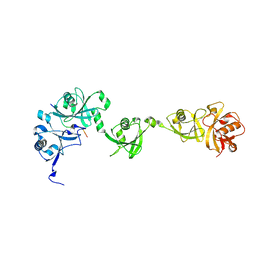 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
4QRP
 
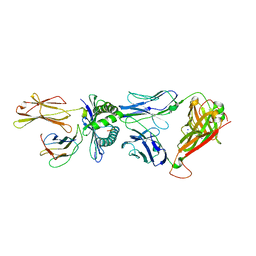 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL and DD31 TCR | | Descriptor: | Beta-2-microglobulin, DD31 TCR alpha chain, DD31 TCR beta chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
5C5A
 
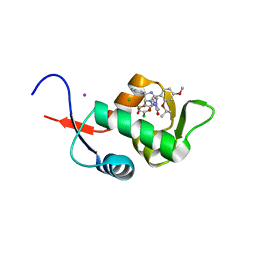 | | Crystal Structure of HDM2 in complex with Nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Orts, J, Waelti, M.A, Marsh, M, Vera, L, Gossert, A.D, Guentert, P, Riek, R. | | Deposit date: | 2015-06-19 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.146 Å) | | Cite: | NMR Molecular Replacement, NMR2
To Be Published
|
|
5C6K
 
 | | Bacteriophage P2 integrase catalytic domain | | Descriptor: | Integrase | | Authors: | Skaar, K, Claesson, M, Odegrip, R, Eriksson, J, Hogbom, M, Haggard-Ljungquist, E, Stenmark, P. | | Deposit date: | 2015-06-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the bacteriophage P2 integrase catalytic domain.
Febs Lett., 589, 2015
|
|
5C6J
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
5MTL
 
 | | Crystal structure of an amyloidogenic light chain | | Descriptor: | light chain dimer,IGL@ protein,IGL@ protein | | Authors: | Oberti, L, Rognoni, P, Russo, R, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2017-01-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
5C9G
 
 | | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium | | Descriptor: | D-MALATE, Enoyl-CoA hydratase/isomerase family protein, TETRAETHYLENE GLYCOL | | Authors: | Szlachta, K, Cooper, D.R, Chapman, H.C, Cymbrowski, M.T, Stead, M, Hillerich, B.S, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, Hammonds, J, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative enoyl-CoA hydratase/isomerase family protein from Hyphomonas neptunium
to be published
|
|
4QUQ
 
 | | Crystal structure of stachydrine demethylase in complex with azide | | Descriptor: | AZIDE ION, COBALT HEXAMMINE(III), FE (III) ION, ... | | Authors: | Agarwal, R, Andi, B, Gizzi, A, Bonanno, J.B, Almo, S.C, Orville, A.M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.266 Å) | | Cite: | Tracking photoelectron induced in-crystallo enzyme catalysis
To be Published
|
|
4QYB
 
 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein, disulfide-bridged dimer, from Burkholderia cenocepacia J2315 | | Descriptor: | Uncharacterized protein | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Shuvalova, L, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein, disulfide-bridged dimer, from Burkholderia cenocepacia J2315
To be Published
|
|
5MWJ
 
 | | Structure Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1 | | Descriptor: | DIMETHYL SULFOXIDE, Transducin-like enhancer protein 1, pepide inhibtor | | Authors: | McGrath, S, Tortorici, M, Vidler, L, Drouin, L, Westwood, I, Gimeson, P, Van Montfort, R, Hoelder, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Enabled Discovery of a Stapled Peptide Inhibitor to Target the Oncogenic Transcriptional Repressor TLE1.
Chemistry, 23, 2017
|
|
5CCN
 
 | | Human Cyclophilin D Complexed with Inhibitor | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, POTASSIUM ION, ... | | Authors: | Gibson, R.P, Shore, E, Kershaw, N, Awais, M, Javed, A, Latawiec, D, Pandalaneni, S, Wen, L, Berry, N, O'Neill, P, Sutton, R, Lian, L.Y. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Cyclophilin D Complexed with Inhibitor
To Be Published
|
|
5CD7
 
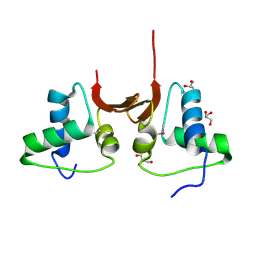 | | Crystal structure of the NTD L199M of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CGS
 
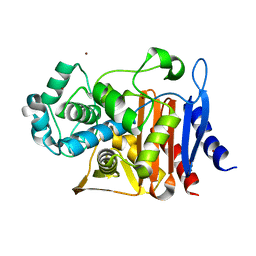 | |
5CH4
 
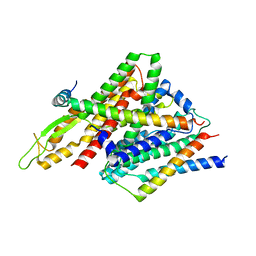 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5MUQ
 
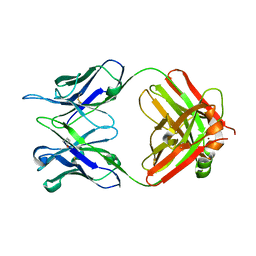 | | Crystal structure of DC8E8 Fab at pH 7.0 containing a Zn atom | | Descriptor: | IMIDAZOLE, ZINC ION, antibody Fab heavy chain, ... | | Authors: | Skrabana, R, Novak, M, Cehlar, O, Kontsekova, E. | | Deposit date: | 2017-01-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of DC8E8 Fab at pH 7.0 containing a Zn atom
To be published
|
|
5MXD
 
 | | BACE-1 IN COMPLEX WITH LIGAND 32397778 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, ~{N},~{N}-dimethyl-2-pyrrolidin-1-yl-quinazolin-4-amine | | Authors: | Alexander, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Human Beta Secretase 1 In Complex With Ligand 32397778
to be published
|
|
5CHU
 
 | | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with sulfate | | Descriptor: | ACETATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Lefurgy, S, Almo, S.C. | | Deposit date: | 2015-07-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CRYSTAL STRUCTURE OF Fox-4 cephamycinase complexed with sulfate
To Be Published
|
|
5M9D
 
 | |
5CLM
 
 | | 1,4-Oxazine BACE1 inhibitors | | Descriptor: | Beta-secretase 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Rombouts, F, Tresadern, G, Delgado, O, Martinez Lamenca, C, Van Gool, M, Garcia-Molina, A, Alonso De Diego, S, Oehlrich, D, Prokopcova, H, Alonso, J.M, Austin, N, Borghys, H, Van Brandt, S, Surkyn, M, De Cleyn, M, Vos, A, Alexander, R, MacDonald, G, Moechars, D, Trabanco, A, Gijsen, H. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-30 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 1,4-Oxazine beta-Secretase 1 (BACE1) Inhibitors: From Hit Generation to Orally Bioavailable Brain Penetrant Leads.
J.Med.Chem., 58, 2015
|
|
5CLS
 
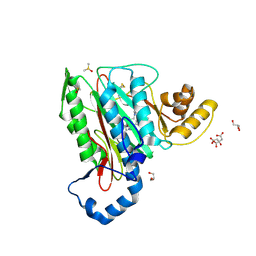 | | Structure of human methionine aminopeptidase-2 complexed with spiroepoxytriazole inhibitor (+)-31a | | Descriptor: | (4R,7R)-7-hydroxy-1-(4-methoxybenzyl)-7-methyl-4,5,6,7-tetrahydro-1H-benzotriazol-4-yl propan-2-ylcarbamate, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Janowski, R, Miller, A.K, Niessing, D. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spiroepoxytriazoles Are Fumagillin-like Irreversible Inhibitors of MetAP2 with Potent Cellular Activity.
Acs Chem.Biol., 11, 2016
|
|
2BML
 
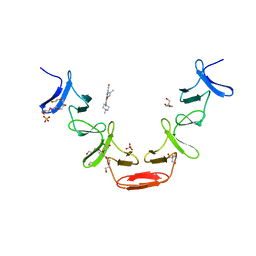 | | Ofloxacin-like antibiotics inhibit pneumococcal cell wall degrading virulence factors | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AUTOLYSIN, DEXTROFLOXACINE, ... | | Authors: | Fernandez-Tornero, C, Gimenez-Gallego, G, Romero, A, Garcia, E, Pascual-Teresa, B.D, Lopez, R. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ofloxacin-Like Antibiotics Inhibit Pneumococcal Cell Wall-Degrading Virulence Factors
J.Biol.Chem., 280, 2005
|
|
5COA
 
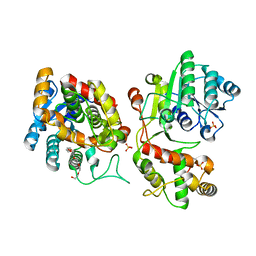 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | Descriptor: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | Authors: | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5N19
 
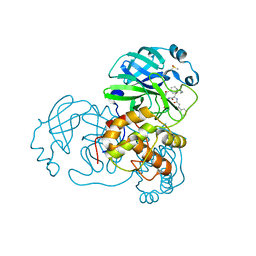 | |
5CPC
 
 | |
