3MJX
 
 | | Crystal structure of myosin-2 motor domain in complex with ADP-Metavanadate and blebbistatin | | Descriptor: | (-)-1-PHENYL-1,2,3,4-TETRAHYDRO-4-HYDROXYPYRROLO[2,3-B]-7-METHYLQUINOLIN-4-ONE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Fedorov, R, Baruch, P, Bauer, S, Manstein, D.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of pentabromopseudilin inhibition of myosin motor activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8E39
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3B
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
7UAW
 
 | | Structure of Clostridium botulinum prophage Tad1 in complex with 1''-2' gcADPR | | Descriptor: | (1S,3R,4R,6R,9S,11R,14R,15S,16R,18R)-4-(6-amino-9H-purin-9-yl)-9,11,15,16,18-pentahydroxy-2,5,8,10,12,17-hexaoxa-9lambda~5~,11lambda~5~-diphosphatricyclo[12.2.1.1~3,6~]octadecane-9,11-dione, ABC transporter ATPase | | Authors: | Lu, A, Leavitt, A, Yirmiya, E, Amitai, G, Garb, J, Morehouse, B.R, Hobbs, S.J, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2022-03-14 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Viruses inhibit TIR gcADPR signalling to overcome bacterial defence.
Nature, 611, 2022
|
|
4WT7
 
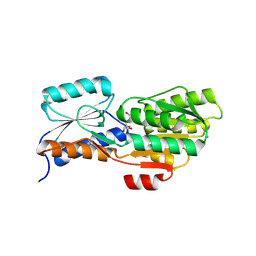 | | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol | | Descriptor: | ABC transporter substrate binding protein (Ribose), CHLORIDE ION, D-allitol | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-10-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an ABC transporter solute binding protein (IPR025997) from Agrobacterium vitis (Avi_5165, Target EFI-511223) with bound allitol
To be published
|
|
8RSB
 
 | | p97 (VCP) mutant - F539A ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) mutant - F539A ADP state
To Be Published
|
|
8E2X
 
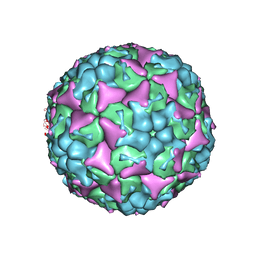 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3C
 
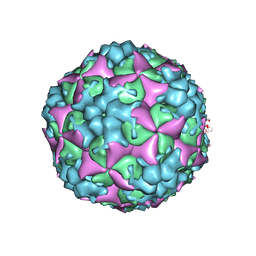 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
1GHX
 
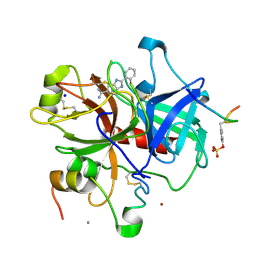 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-HYDROXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GI7
 
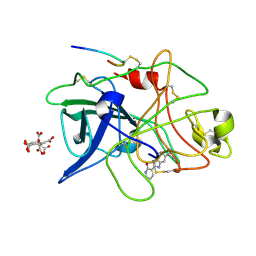 | | A NOVEL SERINE PROTEASE INHIBITION MOTIF INVOLVING A MULTI-CENTERED SHORT HYDROGEN BONDING NETWORK AT THE ACTIVE SITE | | Descriptor: | 2-(2-OXO-1,2-DIHYDRO-PYRIDIN-3-YL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Elrod, K, Luong, C, Rice, M, Mackman, R.L, Sprengeler, P.A, Spencer, J, Hatayte, J, Janc, J, Link, J, Litvak, J, Rai, R, Rice, K, Sideris, S, Verner, E, Young, W. | | Deposit date: | 2001-01-22 | | Release date: | 2002-01-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A novel serine protease inhibition motif involving a multi-centered short hydrogen bonding network at the active site.
J.Mol.Biol., 307, 2001
|
|
1GCI
 
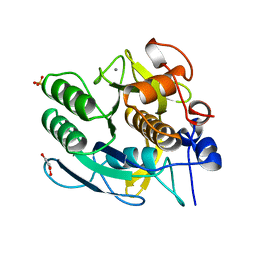 | |
6Q9F
 
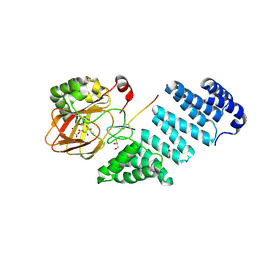 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H679A in complex with Mn, NOG and Factor X peptide fragment (39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, GLYCEROL, ... | | Authors: | Chowdhury, R, Pfeffer, I, Schofield, C.J. | | Deposit date: | 2018-12-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Aspartyl/Asparaginyl beta-hydroxylase (AspH) H679A in complex with Mn, NOG and Factor X peptide fragment (39mer-4Ser)
To Be Published
|
|
8E38
 
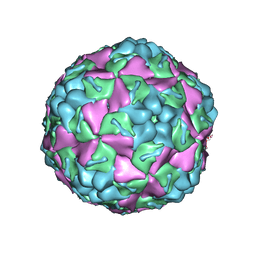 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
6Q9I
 
 | |
8E3A
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
1BRD
 
 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
8RS9
 
 | | p97 (VCP) double mutant - F266A F539A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) double mutant - F266A F539A
To Be Published
|
|
8E2Y
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E31
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
4JVQ
 
 | | Crystal structure of hcv ns5b polymerase in complex with compound 9 | | Descriptor: | 5-{4-[(4-methoxybenzoyl)amino]phenoxy}-2-{[(trans-4-methylcyclohexyl)carbonyl](propan-2-yl)amino}benzoic acid, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-26 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enantiomeric Atropisomers Inhibit HCV Polymerase and/or HIV Matrix: Characterizing Hindered Bond Rotations and Target Selectivity.
J.Med.Chem., 57, 2014
|
|
4JGO
 
 | | The crystal structure of sporulation kinase d sensor domain from Bacillus subtilis subsp. | | Descriptor: | GLYCEROL, PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
8EDD
 
 | | Staphylococcus aureus endonuclease IV Y33F mutant | | Descriptor: | CHLORIDE ION, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Saper, M.A, Kirillov, S, Isupov, M.N, Wiener, R, Rouvinski, A. | | Deposit date: | 2022-09-04 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
To Be Published
|
|
3QSZ
 
 | | Crystal Structure of the STAR-related lipid transfer protein (fragment 25-204) from Xanthomonas axonopodis at the resolution 2.4A, Northeast Structural Genomics Consortium Target XaR342 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, STAR-related lipid transfer protein, ... | | Authors: | Kuzin, A.P, Su, M, Vorobiev, S.M, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Northeast Structural Genomics Consortium Target XaR342
To be Published
|
|
6I9M
 
 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and R-2-hydroxyglutarate | | Descriptor: | (2R)-2-hydroxypentanedioic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2018-11-23 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
4YLI
 
 | | CL-K1 trimer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collectin-11, ... | | Authors: | Wallis, R, Girija, U.V, Gingras, A.R, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis of sugar recognition by collectin-K1 and the effects of mutations associated with 3MC syndrome.
Bmc Biol., 13, 2015
|
|
