7WTL
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Dis-D | | Descriptor: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Cheng, J, La Venuta, G, Lau, B, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In vitro structural maturation of an early stage pre-40S particle coupled with U3 snoRNA release and central pseudoknot formation.
Nucleic Acids Res., 50, 2022
|
|
7WTT
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A1 (with CK1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTP
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (with Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
2XIO
 
 | | Structure of putative deoxyribonuclease TATDN1 isoform a | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, PUTATIVE DEOXYRIBONUCLEASE TATDN1, ... | | Authors: | Muniz, J.R.C, Cocking, R, Puranik, S, Krojer, T, Raynor, J, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of Putative Deoxyribonuclease Tatdn1 Isoform A
To be Published
|
|
2XYH
 
 | | Caspase-3:CAS60254719 | | Descriptor: | 5-CHLORO-4-OXOPENTANOIC ACID, CASPASE-3 SUBUNIT P12, CASPASE-3 SUBUNIT P17 | | Authors: | Ganesan, R, Jelakovic, S, Grutter, M.G, Mittl, P.R. | | Deposit date: | 2010-11-17 | | Release date: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | In Silico Identification and Crystal Structure Validation of Caspase-3 Inhibitors without a P1 Aspartic Acid Moiety.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
7WTX
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTZ
 
 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-B2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
2XO0
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to 24-diamino-1,3,5- triazine identified by virtual screening | | Descriptor: | 1,3,5-TRIAZINE-2,4-DIAMINE, ACETATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
2KPW
 
 | | NMR solution structure of Lamin-B1 protein from Homo sapiens: Northeast Structural Genomics Consortium MEGA target, HR5546A (439-549) | | Descriptor: | Lamin-B1 | | Authors: | Swapna, G.V.T, Ciccosanti, C.L, Belote, R, Hamilton, K, Acton, T, Huang, Y, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Lamin-B1 protein from Homo sapiens: Northeast Structural Genomics Consortium target, HR5546A (439-549)
To be Published
|
|
5O2T
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
2K52
 
 | | Structure of uncharacterized protein MJ1198 from Methanocaldococcus jannaschii. Northeast Structural Genomics Target MjR117B | | Descriptor: | Uncharacterized protein MJ1198 | | Authors: | Rossi, P, Maglaqui, M, Foote, E.L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Swapna, G, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-24 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of uncharacterized protein MJ1198 from
Methanocaldococcus jannaschii. Northeast Structural Genomics Target MjR117B
To be Published
|
|
3K4W
 
 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
2Y61
 
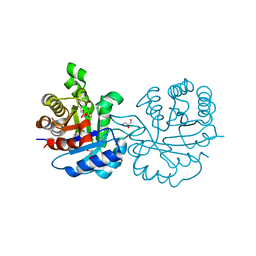 | | Crystal structure of Leishmanial E65Q-TIM complexed with S-Glycidol phosphate | | Descriptor: | GLYCEROL, SN-GLYCEROL-1-PHOSPHATE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Venkatesan, R, Alahuhta, M, Pihko, P.M, Wierenga, R.K. | | Deposit date: | 2011-01-19 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | High resolution crystal structures of triosephosphate isomerase complexed with its suicide inhibitors: the conformational flexibility of the catalytic glutamate in its closed, liganded active site.
Protein Sci., 20, 2011
|
|
7B0B
 
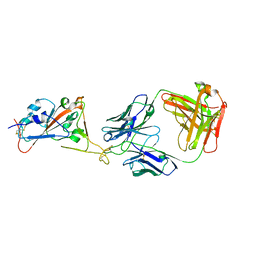 | | Fab HbnC3t1p1_C6 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, IGK@ protein, ... | | Authors: | Borenstein-Katz, A, Diskin, R. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Somatic hypermutation introduces bystander mutations that prepare SARS-CoV-2 antibodies for emerging variants.
Immunity, 56, 2023
|
|
2XOC
 
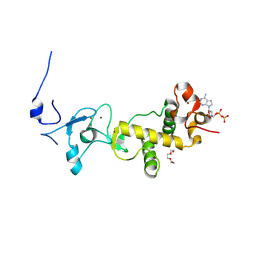 | | C-terminal cysteine-rich domain of human CHFR bound to mADPr | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ... | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
2XGF
 
 | | Structure of the bacteriophage T4 long tail fibre needle-shaped receptor-binding tip | | Descriptor: | CARBONATE ION, FE (II) ION, LONG TAIL FIBER PROTEIN P37 | | Authors: | Bartual, S.G, Otero, J.M, Garcia-Doval, C, Llamas-Saiz, A.L, Kahn, R, Fox, G.C, van Raaij, M.J. | | Deposit date: | 2010-06-03 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the bacteriophage T4 long tail fiber receptor-binding tip.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2KPO
 
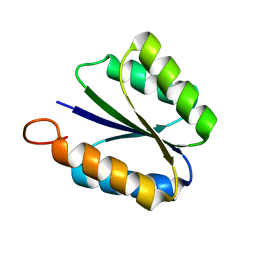 | | Solution NMR structure of de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium target OR16 | | Descriptor: | rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Hamilton, K, Ciccosanti, C, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-17 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of denovo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium target OR16
To be Published
|
|
2VOC
 
 | | THIOREDOXIN A ACTIVE SITE MUTANTS FORM MIXED DISULFIDE DIMERS THAT RESEMBLE ENZYME-SUBSTRATE REACTION INTERMEDIATE | | Descriptor: | DI(HYDROXYETHYL)ETHER, THIOREDOXIN | | Authors: | Kouwen, T.R.H.M, Andrell, J, Schrijver, R, Dubois, J.Y.F, Maher, M.J, Iwata, S, Carpenter, E.P, van Dijl, J.M. | | Deposit date: | 2008-02-13 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates.
J. Mol. Biol., 379, 2008
|
|
2VPI
 
 | | Human GMP synthetase - glutaminase domain | | Descriptor: | GMP SYNTHASE | | Authors: | Welin, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Van Der Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human Gmp Synthetase - Glutaminase Domain
To be Published
|
|
7B0W
 
 | | Crystal structure of the E. coli type 1 pilus assembly inhibitor FimI bound to FimC | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, FORMIC ACID, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
2VVT
 
 | | Glutamate Racemase (MurI) from E. faecalis in complex with a 9-Benzyl Purine inhibitor | | Descriptor: | 2-butoxy-9-(2,6-difluorobenzyl)-N-(2-morpholin-4-ylethyl)-9H-purin-6-amine, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Geng, B, Breault, G, Comita-Prevoir, J, Petrichko, R, Eyermann, C, Lundqvist, T, Doig, P, Gorseth, E, Noonan, B. | | Deposit date: | 2008-06-11 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Exploring 9-Benzyl Purines as Inhibitors of Glutamate Racemase (Muri) in Gram-Positive Bacteria.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7B0X
 
 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
2VWO
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3S,4S)-4-FLUORO- 1-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VXQ
 
 | | Crystal structure of the major grass pollen allergen Phl p 2 in complex with its specific IgE-Fab | | Descriptor: | FAB, POLLEN ALLERGEN PHL P 2 | | Authors: | Padavattan, S, Flicker, S, Schirmer, T, Madritsch, C, Randow, S, Reese, G, Vieths, S, Lupinek, C, Ebner, C, Valenta, R, Markovic-Housley, Z. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Ige Recognition of a Conformational Epitope of the Major Respiratory Allergen Phl P 2 as Revealed by X-Ray Crystallography.
J.Immunol., 182, 2009
|
|
2VWM
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | (4R)-4-{[(5-chlorothiophen-2-yl)carbonyl]amino}-N-(cyclopropylmethyl)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-L-prolinamide, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
