2LUL
 
 | | Solution NMR Structure of PH Domain of Tyrosine-protein kinase Tec from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR3504C | | Descriptor: | Tyrosine-protein kinase Tec, ZINC ION | | Authors: | Liu, G, Xiao, R, Janjua, H, Hamilton, K, Shastry, R, Kohan, E, Acton, T.B, Everett, J.K, Lee, H, Pederson, K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PH Domain of Tyrosine-protein kinase Tec from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR3504C
To be Published
|
|
2M6Q
 
 | | Refined Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Strucutral Genomics Consortium Target ZR18 | | Descriptor: | SAV1430 | | Authors: | Baran, M.C, Aramini, J.M, Huang, Y.J, Xiao, R, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PDBStat: a universal restraint converter and restraint analysis software package for protein NMR.
J.Biomol.Nmr, 56, 2013
|
|
6BFL
 
 | | Caspase-3 Mutant- D9A,D28A,T245D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3 | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6BFO
 
 | | Caspase-3 Mutant- T245D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6B04
 
 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6AY4
 
 | | Naegleria fowleri CYP51-fluconazole complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, CYP51, ... | | Authors: | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
2RVO
 
 | | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish | | Descriptor: | RNA (34-MER) | | Authors: | Otsu, M, Norose, N, Arai, N, Terao, R, Kajikawa, M, Okada, N, Kawai, G. | | Deposit date: | 2016-02-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a reverse transcriptase recognition site of a LINE RNA from zebrafish.
J. Biochem., 162, 2017
|
|
2JHR
 
 | | Crystal structure of myosin-2 motor domain in complex with ADP- metavanadate and pentabromopseudilin | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN, ... | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3ESH
 
 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | Descriptor: | Protein similar to metal-dependent hydrolase | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
6BG4
 
 | | Caspase-3 Mutant- T152D | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, AZIDE ION, CHLORIDE ION, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-27 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
2LV2
 
 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
2JJ9
 
 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
4LNS
 
 | |
6BD2
 
 | |
4C2E
 
 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
3F2Z
 
 | | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B | | Descriptor: | uncharacterized protein BF3579 | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, Foote, E.L, Ciccosanti, C, Lee, D, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B
To be Published
|
|
4C2D
 
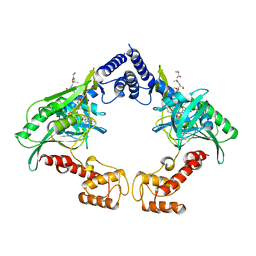 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4LMF
 
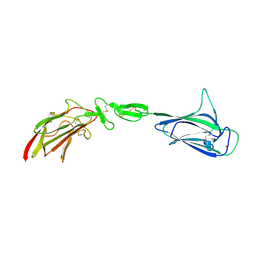 | | C1s CUB1-EGF-CUB2 | | Descriptor: | CALCIUM ION, Complement C1s subcomponent heavy chain, SODIUM ION | | Authors: | Wallis, R, Venkatraman Girija, U, Moody, P.C.E, Marshall, J.E. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-07 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.921 Å) | | Cite: | Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6BCC
 
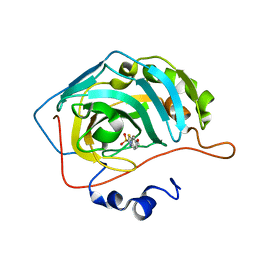 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with ethoxzolamide | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BFK
 
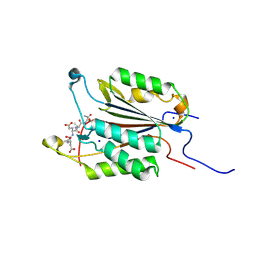 | | Caspase-3 Mutant- T245A | | Descriptor: | AC-ASP-GLU-VAL-ASP-CMK, Caspase-3, SODIUM ION | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
6B73
 
 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
6B9H
 
 | |
6BC9
 
 | | Joint X-ray/neutron structure of human carbonic anhydrase II in complex with dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Kovalevsky, A, McKenna, R, Aggarwal, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | "To Be or Not to Be" Protonated: Atomic Details of Human Carbonic Anhydrase-Clinical Drug Complexes by Neutron Crystallography and Simulation.
Structure, 26, 2018
|
|
6BD1
 
 | |
6BCY
 
 | |
