5AXI
 
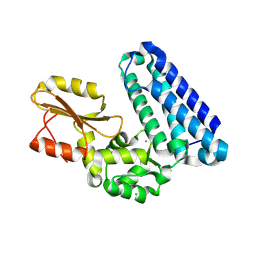 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
7ZCA
 
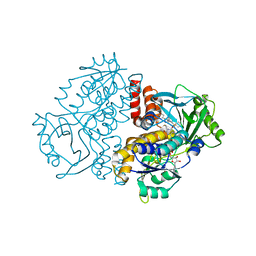 | | Crystal structure of the F191M/F201A variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with benzyl phenyl sulfoxide | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Putative dehydrogenase/oxygenase subunit (Flavoprotein), ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-26 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7Z99
 
 | |
5AZ9
 
 | |
4Q51
 
 | | Crystal structure of a putative molybdenum cofactor biosynthesis protein F from Burkholderia cenocepacia J2315 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Minasov, G, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative molybdenum cofactor biosynthesis protein F from Burkholderia cenocepacia J2315
To be Published
|
|
4QBY
 
 | | yCP in complex with BOC-ALA-ALA-ALA-CHO | | Descriptor: | BOC-ALA-ALA-ALA-CHO, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Arciniega, M, Beck, P, Lange, O, Groll, M, Huber, R. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7Z97
 
 | | Crystal structure of the F191M variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with 6-bromoindole | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5AQV
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | (1R,2S,3R,5R)-3-((5-(benzyloxy)quinazolin-4-yl)amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
4PW4
 
 | | Crystal structure of Aminopeptidase N in complex with phosphonic acid analogue of homophenylalanine L-(R)-hPheP | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Mulligan, R, Vassiliou, S, Berlicki, L, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-18 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with phosphonic analogs of homophenylalanine
TO BE PUBLISHED
|
|
5AQJ
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9H-purine-6,8-diamine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7K98
 
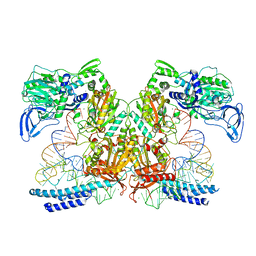 | | Preaminoacylation complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine (F-AMS) | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
5LFD
 
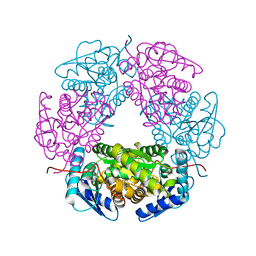 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ramazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-01 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
5LG3
 
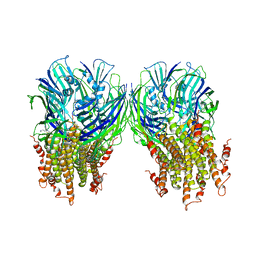 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.567 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7JPM
 
 | |
5LCC
 
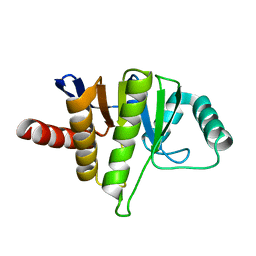 | | Oceanobacillus iheyensis macrodomain mutant D40A | | Descriptor: | MACROD-TYPE MACRODOMAIN | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5AQF
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AQS
 
 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, GLYCEROL, HEAT SHOCK COGNATE 71 KDA PROTEIN, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
5AP1
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 6-{[3-(cyanomethoxy)-4-(1-methyl-1H-pyrazol-4-yl)phenyl]amino}-2-(cyclohexylamino)pyridine-3-carbonitrile, DUAL SPECIFICITY PROTEIN KINASE TTK, GLYCEROL, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
5LJD
 
 | | Crystal structure of holo human CRBP1/K40L mutant | | Descriptor: | RETINOL, Retinol-binding protein 1, SODIUM ION | | Authors: | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | Deposit date: | 2016-07-18 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5APE
 
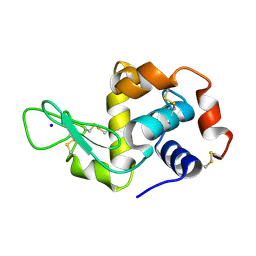 | | Hen Egg White Lysozyme reference dataset odd frames | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
5LKA
 
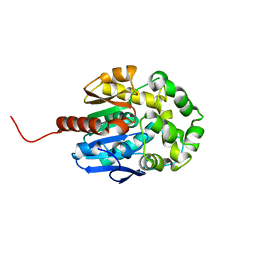 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
4Q33
 
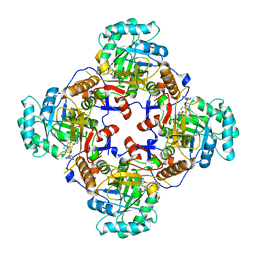 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110 | | Descriptor: | 4-[(1R)-1-[1-(4-chlorophenyl)-1,2,3-triazol-4-yl]ethoxy]-1-oxidanyl-quinoline, ACETIC ACID, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Gu, M, Zhang, M, Mandapati, K, Gollapalli, D.R, Gorla, S.K, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and A110
TO BE PUBLISHED
|
|
3R8L
 
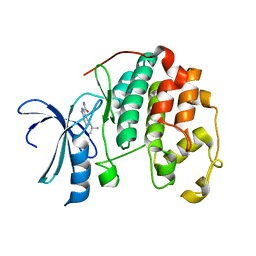 | | CDK2 in complex with inhibitor L3-4 | | Descriptor: | (5R)-5-tert-butyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
7K1D
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM_77291 | | Descriptor: | (3R)-3-{4-[(3R)-4-(3,4-difluorobenzene-1-carbonyl)morpholin-3-yl]-1H-1,2,3-triazol-1-yl}-N-hydroxy-4-(naphthalen-2-yl)butanamide, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Bosc, D, Tang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 2
To Be Published
|
|
