7QFH
 
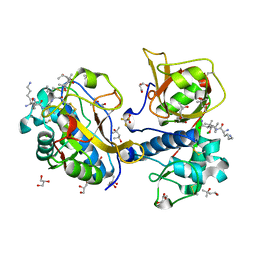 | | Peptide AYFKKVL in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AMINO GROUP, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7QHJ
 
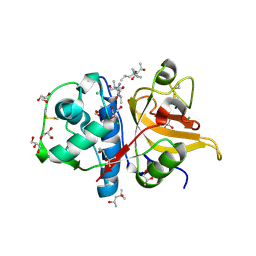 | | Peptide GAKSAA in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
8E3D
 
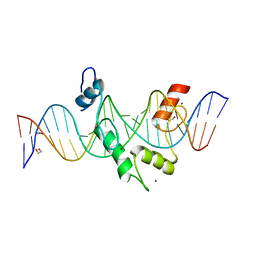 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#11) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*AP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*AP*T)-3'), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
1FVR
 
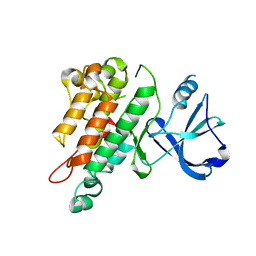 | | TIE2 KINASE DOMAIN | | Descriptor: | TYROSINE-PROTEIN KINASE TIE-2 | | Authors: | Shewchuk, L.M, Hassell, A.M, Ellis, B, Holmes, W.D, Davis, R, Horne, E.L, Kadwell, S.H, McKee, D.D, Moore, J.T. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Tie2 RTK domain: self-inhibition by the nucleotide binding loop, activation loop, and C-terminal tail.
Structure Fold.Des., 8, 2000
|
|
8E76
 
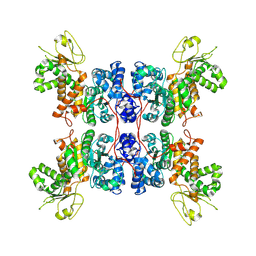 | | Cryo-EM structure of Apo form ME3 | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8E8O
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
7QHK
 
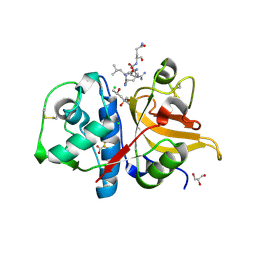 | | Peptide QLRQQE in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
7QLR
 
 | |
1FV1
 
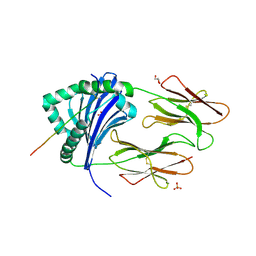 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
8E3E
 
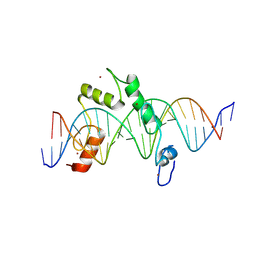 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#10) | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*A)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8E78
 
 | | Cryo-EM structure of human ME3 in the presence of citrate | | Descriptor: | NADP-dependent malic enzyme, mitochondrial | | Authors: | Yu, X, Grell, T.A.J, Shaffer, P.L, Steele, R, Sharma, S, Thompson, A.A, Tresadern, G, Ortiz-Meoz, R.F, Mason, M, Gomez-Tamayo, J.C, Riley, D, Wagner, M.V, Wadia, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
4WPI
 
 | |
1FZU
 
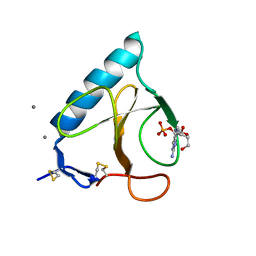 | | RNAse T1 V78A mutant | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Vos, S, Loris, R, Steyaert, J. | | Deposit date: | 2000-10-04 | | Release date: | 2000-10-25 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrophobic core manipulations in ribonuclease T1.
Biochemistry, 40, 2001
|
|
1G02
 
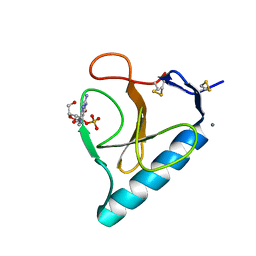 | | Ribonuclease T1 V16S mutant | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, GUANYL-SPECIFIC RIBONUCLEASE T1 | | Authors: | De Vos, S, Loris, R, Steyaert, J. | | Deposit date: | 2000-10-05 | | Release date: | 2000-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Hydrophobic core manipulations in ribonuclease T1.
Biochemistry, 40, 2001
|
|
7QKA
 
 | | Crystal structure of SARS-CoV-2 Main Protease in complex with covalently bound GC376 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKC
 
 | | Crystal structure of human Cathepsin L after incubation with Sulfo-Calpeptin | | Descriptor: | Calpeptin, Cathepsin L, DI(HYDROXYETHYL)ETHER | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
6PV8
 
 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with AT-1001 | | Descriptor: | (3-endo)-N-(2-bromophenyl)-9-methyl-9-azabicyclo[3.3.1]nonan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
7QNS
 
 | | Peptide VYEKKP in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide VYEKKP in complex with human cathepsin V C25S mutant
To Be Published
|
|
1G63
 
 | | PEPTIDYL-CYSTEINE DECARBOXYLASE EPID | | Descriptor: | EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbac, S. | | Deposit date: | 2000-11-03 | | Release date: | 2001-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1G71
 
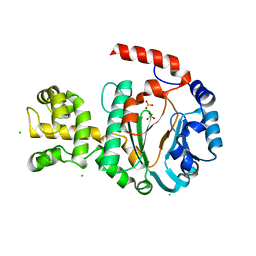 | | CRYSTAL STRUCTURE OF PYROCOCCUS FURIOSUS DNA PRIMASE | | Descriptor: | CHLORIDE ION, DNA PRIMASE, SULFATE ION, ... | | Authors: | Augustin, M.A, Huber, R, Kaiser, J.T. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a DNA-dependent RNA polymerase (DNA primase).
Nat.Struct.Biol., 8, 2001
|
|
8DPK
 
 | |
6PS3
 
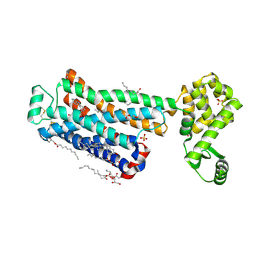 | | XFEL beta2 AR structure by ligand exchange from Timolol to Carvedilol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(8H-CARBAZOL-4-YLOXY)-3-[2-(2-METHOXYPHENOXY)ETHYLAMINO]PROPAN-2-OL, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8SME
 
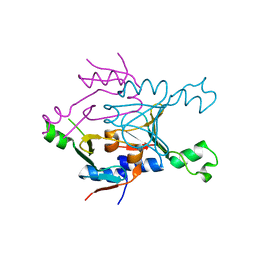 | | Structure of SPO1 phage Tad2 in apo state | | Descriptor: | Gp34.65 | | Authors: | Lu, A, Yirmiya, E, Leavitt, A, Avraham, C, Osterman, I, Garb, J, Antine, S.P, Mooney, S.E, Hobbs, S.J, Amitai, G, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Phages overcome bacterial immunity via diverse anti-defence proteins.
Nature, 625, 2024
|
|
1G84
 
 | | THE SOLUTION STRUCTURE OF THE C EPSILON2 DOMAIN FROM IGE | | Descriptor: | IMMUNOGLOBULIN E | | Authors: | McDonnell, J.M, Cowburn, D, Gould, H.J, Sutton, B.J, Calvert, R, Beavil, R.E, Beavil, A.J, Henry, A.J. | | Deposit date: | 2000-11-16 | | Release date: | 2001-05-16 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha.
Nat.Struct.Biol., 8, 2001
|
|
8DWL
 
 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
