3KU6
 
 | | Crystal structure of a H2N2 influenza virus hemagglutinin, 226L/228G | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-19 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic.
J.Virol., 84, 2010
|
|
3I3S
 
 | | Crystal Structure of H-Ras with Thr50 replaced by Isoleucine | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gremer, L, Dvorsky, R, Merbitz-Zahradnik, T, Wittinghofer, A, Ahmadian, M.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A restricted spectrum of NRAS mutations causes Noonan syndrome.
Nat.Genet., 42, 2010
|
|
5FPL
 
 | | Crystal structure of human JARID1B in complex with CCT363901 | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(2-azanylethyl)pyrazol-1-yl]-3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Yann-Vai, L.B, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Huber, K, Oppermann, U. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 8-Substituted Pyrido[3,4-D]Pyrimidin-4(3H)-One Derivatives as Potent, Cell Permeable, Kdm4 (Jmjd2) and Kdm5 (Jarid1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2ZIO
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AlocLys-AMP and PNP | | Descriptor: | 5'-O-[(S)-({(2S)-2-amino-6-[(propoxycarbonyl)amino]hexanoyl}oxy)(hydroxy)phosphoryl]adenosine, IMIDODIPHOSPHORIC ACID, Pyrrolysyl-tRNA synthetase | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
5HEG
 
 | |
3LE7
 
 | | Crystal structure of PD-L1 from P. dioica in complex with adenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein PD-L1/PD-L2 | | Authors: | Ruggiero, A, Berisio, R. | | Deposit date: | 2010-01-14 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The role of the glycan moiety on the structure-function relationships of PD-L1, type 1 ribosome-inactivating protein from P. dioica leaves
Mol Biosyst, 6, 2010
|
|
3KMS
 
 | | G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA trigonal structure | | Descriptor: | 3D polymerase, MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*GP*GP*CP*C)-3'), ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
1QP9
 
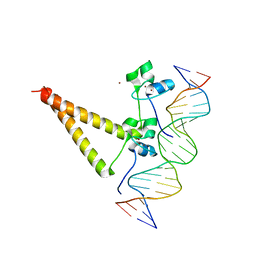 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
3KXH
 
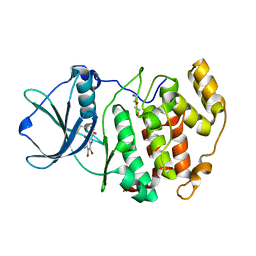 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor (2-dymethylammino-4,5,6,7-tetrabromobenzoimidazol-1yl-acetic acid (K66) | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, [4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzimidazol-1-yl]acetic acid | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3KU3
 
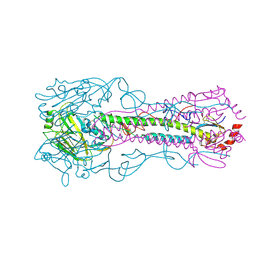 | | Crystal structure of a H2N2 influenza virus hemagglutinin, avian like | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic.
J.Virol., 84, 2010
|
|
3KOA
 
 | | M296I mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA and GTP | | Descriptor: | 3D polymerase, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-13 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
3KXN
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor tetraiodobenzimidazole (K88) | | Descriptor: | 4,5,6,7-tetraiodo-1H-benzimidazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3KNA
 
 | | M296I mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA | | Descriptor: | 3D polymerase, MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*GP*GP*CP*C)-3'), ... | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-12 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
2ZE1
 
 | | X-ray structure of Bace-1 in complex with compound 6g | | Descriptor: | 3-bromo-N-[4-[1-(2-carbamimidamido-2-oxo-ethyl)-5-phenyl-pyrrol-2-yl]phenyl]benzamide, Beta-secretase 1 | | Authors: | Chopra, R, Olland, A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3LC5
 
 | | Selective Benzothiophine Inhibitors of Factor IXa | | Descriptor: | 1-{4-[(R)-phenyl(3-phenyl-1,2,4-oxadiazol-5-yl)methoxy]-1-benzothiophen-2-yl}methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3AY5
 
 | |
1R4C
 
 | |
3CM8
 
 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
1ZTA
 
 | | THE SOLUTION STRUCTURE OF A LEUCINE-ZIPPER MOTIF PEPTIDE | | Descriptor: | LEUCINE ZIPPER MONOMER | | Authors: | Saudek, V, Pastore, A, Castiglione Morelli, M.A, Frank, R, Gausepohl, H, Gibson, T. | | Deposit date: | 1990-10-11 | | Release date: | 1993-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a leucine-zipper motif peptide.
Protein Eng., 4, 1991
|
|
2B5S
 
 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, LAURIC ACID, Non-specific lipid transfer protein, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, R.J, Zanotti, G. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
3D24
 
 | | Crystal structure of ligand-binding domain of estrogen-related receptor alpha (ERRalpha) in complex with the peroxisome proliferators-activated receptor coactivator-1alpha box3 peptide (PGC-1alpha) | | Descriptor: | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR1 | | Authors: | Moras, D, Greschik, H, Flaig, R, Sato, Y, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Communication between the ERR{alpha} Homodimer Interface and the PGC-1{alpha} Binding Surface via the Helix 8-9 Loop.
J.Biol.Chem., 283, 2008
|
|
6UM2
 
 | | Structure of M-6-P/IGFII Receptor and IGFII complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, Insulin-like growth factor II | | Authors: | Wang, R, Qi, X, Li, X. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments
Sci Adv, 6, 2020
|
|
6UN9
 
 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
5I5A
 
 | | quasi racemic structure of allo-Ile7-ShK and D-ShK | | Descriptor: | D-MALATE, D-ShK, GLYCEROL, ... | | Authors: | Dang, B, Kubota, T, Mandal, K, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2D9Q
 
 | | Crystal Structure of the Human GCSF-Receptor Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CSF3, Granulocyte colony-stimulating factor receptor | | Authors: | Tamada, T, Kuroki, R. | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
