5H4E
 
 | |
5GXP
 
 | |
6TVZ
 
 | | Structure of a psychrophilic CCA-adding enzyme crystallized in the XtalController device | | 分子名称: | ACETATE ION, CCA-adding enzyme, GLYCEROL, ... | | 著者 | de Wijn, R, Rollet, K, Coudray, L, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | 登録日 | 2020-01-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Monitoring the Production of High Diffraction-Quality Crystals of Two Enzymes in Real Time Using In Situ Dynamic Light Scattering
Crystals, 2020
|
|
5H23
 
 | | Crystal structure of Chikungunya virus capsid protein | | 分子名称: | 1,2-ETHANEDIOL, Capsid Protein, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sharma, R, Kesari, P, Tomar, S, Kumar, P. | | 登録日 | 2016-10-14 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-function insights into chikungunya virus capsid protein: Small molecules targeting capsid hydrophobic pocket.
Virology, 515, 2018
|
|
6U42
 
 | | Natively decorated ciliary doublet microtubule | | 分子名称: | DC1, DC2, DC3, ... | | 著者 | Ma, M, Stoyanova, M, Rademacher, G, Dutcher, S.K, Brown, A, Zhang, R. | | 登録日 | 2019-08-22 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the Decorated Ciliary Doublet Microtubule.
Cell, 179, 2019
|
|
5H45
 
 | | Crystal structure of the C-terminal Lon protease-like domain of Thermus thermophilus RadA/Sms | | 分子名称: | DNA repair protein RadA | | 著者 | Inoue, M, Fukui, K, Fujii, Y, Nakagawa, N, Yano, T, Kuramitsu, S, Masui, R. | | 登録日 | 2016-10-30 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The Lon protease-like domain in the bacterial RecA paralog RadA is required for DNA binding and repair.
J. Biol. Chem., 292, 2017
|
|
6TWE
 
 | |
3BT0
 
 | | Crystal structure of transthyretin variant V20S | | 分子名称: | Transthyretin | | 著者 | Zanotti, G, Folli, C, Cendron, L, Gliubich, F, Negro, A, Berni, R. | | 登録日 | 2007-12-27 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
5GAJ
 
 | | Solution NMR structure of De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258 | | 分子名称: | DE NOVO DESIGNED PROTEIN OR258 | | 著者 | Liu, G, Castelllanos, J, Koga, R, Koga, N, Xiao, R, Pederson, K, Janjua, H, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-12-01 | | 公開日 | 2016-01-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure De novo designed PLOOP2X3_50 fold protein, Northeast Structural Genomics Consortium (NESG) target OR258
To Be Published
|
|
5G5T
 
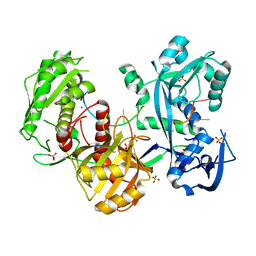 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | 分子名称: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | 著者 | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | 登録日 | 2016-06-03 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
5G1X
 
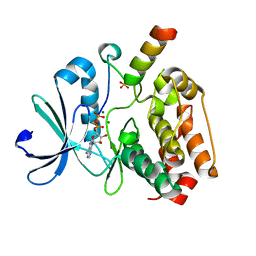 | | Crystal structure of Aurora-A kinase in complex with N-Myc | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, MAGNESIUM ION, ... | | 著者 | Richards, M.W, Burgess, S.G, Bayliss, R. | | 登録日 | 2016-03-31 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5G5W
 
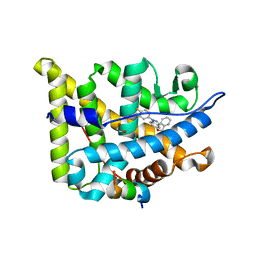 | | Structure guided design and discovery of Indazole ethers as highly potent, non-steroidal Glucocorticoid receptor modulators | | 分子名称: | 1,2-ETHANEDIOL, 2,2,2-trifluoro-N-[(1R,2S)-1-{[1-(4-fluorophenyl)-1H-indazol-5-yl]oxy}-1-phenylpropan-2-yl]acetamide, GLUCOCORTICOID RECEPTOR, ... | | 著者 | Hemmerling, M, Edman, K, Lepisto, M, Eriksson, A, Ivanova, S, Dahmen, J, Rehwinkel, H, Berger, M, Hendrickx, R, Dearman, M, Jellesmark-Jensen, T, Wissler, L, Hansson, T. | | 登録日 | 2016-06-08 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of Indazole Ethers as Novel, Potent, Non-Steroidal Glucocorticoid Receptor Modulators.
Bioorg.Med.Chem.Lett., 26, 2017
|
|
6SP4
 
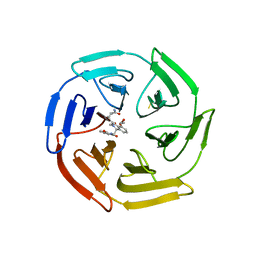 | | KEAP1 IN COMPLEX WITH COMPOUND 23 | | 分子名称: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclobutane-1-carboxamide, Kelch-like ECH-associated protein 1 | | 著者 | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | 登録日 | 2019-08-30 | | 公開日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
5DZC
 
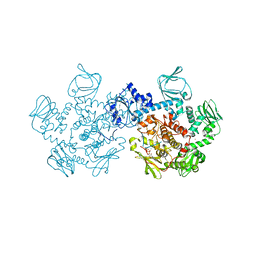 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - AMPPNP bound | | 分子名称: | CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | 著者 | Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, He, H, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | 登録日 | 2015-09-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5E0W
 
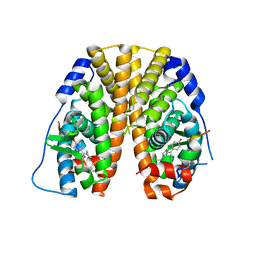 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3S)-3-(4-hydroxyphenyl)cyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-{[(3S)-3-(4-hydroxyphenyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3C5I
 
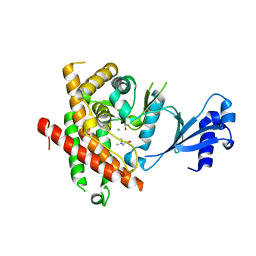 | | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520 | | 分子名称: | CALCIUM ION, CHOLINE ION, Choline kinase, ... | | 著者 | Wernimont, A.K, Hills, T, Lew, J, Wasney, G, Senesterra, G, Kozieradzki, I, Cossar, D, Vedadi, M, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | 登録日 | 2008-01-31 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Plasmodium knowlesi choline kinase, PKH_134520.
To be Published
|
|
4HH1
 
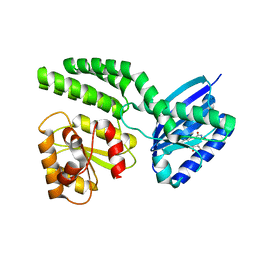 | | Dark-state structure of AppA wild-type without the Cys-rich region from Rb. sphaeroides | | 分子名称: | AppA protein, FLAVIN MONONUCLEOTIDE | | 著者 | Winkler, A, Heintz, U, Lindner, R, Reinstein, J, Shoeman, R, Schlichting, I. | | 登録日 | 2012-10-09 | | 公開日 | 2013-06-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5E3B
 
 | | Structure of macrodomain protein from Streptomyces coelicolor | | 分子名称: | 1,2-ETHANEDIOL, Macrodomain protein, SODIUM ION | | 著者 | Lalic, J, Posavec Marjanovic, M, Perina, D, Sabljic, I, Zaja, R, Plese, B, Imesek, M, Bucca, G, Ahel, M, Cetkovic, H, Luic, M, Mikoc, A, Ahel, I. | | 登録日 | 2015-10-02 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Disruption of Macrodomain Protein SCO6735 Increases Antibiotic Production in Streptomyces coelicolor.
J.Biol.Chem., 291, 2016
|
|
6SU4
 
 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | 著者 | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | 登録日 | 2019-09-12 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SUH
 
 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone, a tolcapone analogue | | 分子名称: | 3-O-methyltolcapone, Transthyretin | | 著者 | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | 登録日 | 2019-09-14 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
6SVI
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x42) | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | 登録日 | 2019-09-18 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVX
 
 | | Reference structure of bovine trypsin (odd frames of crystal x33) | | 分子名称: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | 著者 | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | 登録日 | 2019-09-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-01-29 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SW4
 
 | | The structure of AraP, an arabinose binding protein from Geobacillus stearothermophilus | | 分子名称: | Arabinose binding protein | | 著者 | Lansky, S, Salama, R, Lavid, N, Shoham, Y, Shoham, G. | | 登録日 | 2019-09-19 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.845 Å) | | 主引用文献 | The structure of AraP, an arabinose binding protein from Geobacillus stearothermophilus
To Be Published
|
|
5E7U
 
 | |
6SYM
 
 | |
