2ANT
 
 | | THE 2.6 A STRUCTURE OF ANTITHROMBIN INDICATES A CONFORMATIONAL CHANGE AT THE HEPARIN BINDING SITE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-allopyranose, ANTITHROMBIN | | 著者 | Skinner, R, Abrahams, J.-P, Whisstock, J.C, Lesk, A.M, Carrell, R.W, Wardell, M.R. | | 登録日 | 1997-01-28 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site.
J.Mol.Biol., 266, 1997
|
|
5DOR
 
 | | P2 Integrase catalytic domain in space group P21 | | 分子名称: | Integrase, PHOSPHATE ION, ZINC ION | | 著者 | Skaar, K, Claesson, M, Odegrip, R, Haggard-Ljungquist, E, Hogbom, M. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the bacteriophage P2 integrase catalytic domain.
Febs Lett., 589, 2015
|
|
7L1K
 
 | |
5LVX
 
 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | 分子名称: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | 登録日 | 2016-09-14 | | 公開日 | 2017-10-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
5E1C
 
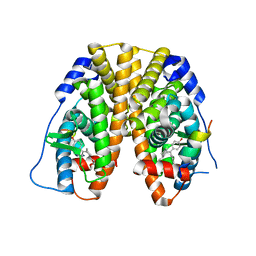 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative dimethyl {(1S)-3-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}propanedioate | | 分子名称: | Estrogen receptor, Nuclear receptor coactivator 2, dimethyl {(1S)-3-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}propanedioate | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DSM
 
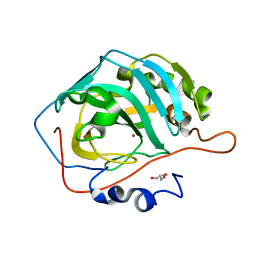 | |
5DSR
 
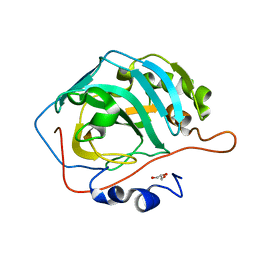 | |
5E21
 
 | | PDZ2 of LNX2 at 277K,single conformer model | | 分子名称: | Ligand of Numb protein X 2 | | 著者 | Hekstra, D.R, White, K.I, Socolich, M.A, Ranganathan, R. | | 登録日 | 2015-09-30 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.011 Å) | | 主引用文献 | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
4U8F
 
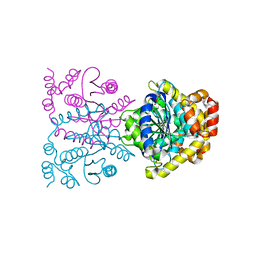 | | Crystal structure of 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase complexed with a tartrate | | 分子名称: | L(+)-TARTARIC ACID, Putative uncharacterized protein gbs1892 | | 著者 | Maruyama, Y, Oiki, S, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2014-08-03 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Metabolic Fate of Unsaturated Glucuronic/Iduronic Acids from Glycosaminoglycans: MOLECULAR IDENTIFICATION AND STRUCTURE DETERMINATION OF STREPTOCOCCAL ISOMERASE AND DEHYDROGENASE.
J.Biol.Chem., 290, 2015
|
|
5M1S
 
 | | Cryo-EM structure of the E. coli replicative DNA polymerase-clamp-exonuclase-theta complex bound to DNA in the editing mode | | 分子名称: | DNA Primer Strand, DNA Template Strand, DNA polymerase III subunit alpha, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2016-10-10 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Self-correcting mismatches during high-fidelity DNA replication.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4U9G
 
 | | Crystal structure of an H-NOX protein from S. oneidensis in the Fe(II)CO ligation state, Q154A/Q155A/K156A mutant | | 分子名称: | CARBON MONOXIDE, NO-binding heme-dependent sensor protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Herzik Jr, M.A, Jonnalagadda, R, Kuriyan, J, Marletta, M.A. | | 登録日 | 2014-08-06 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights into the role of iron-histidine bond cleavage in nitric oxide-induced activation of H-NOX gas sensor proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5DTV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dimethyl-substituted, 3,4-diarylthiophene dioxide core ligand | | 分子名称: | 3,4-bis(4-hydroxy-2-methylphenyl)-1H-1lambda~6~-thiophene-1,1-dione, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-18 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DVN
 
 | | Fc K392D/K409D homodimer | | 分子名称: | Fc-III peptide, Ig gamma-1 chain C region | | 著者 | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | 登録日 | 2015-09-21 | | 公開日 | 2016-03-30 | | 最終更新日 | 2016-07-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
4U10
 
 | |
4U16
 
 | | M3-mT4L receptor bound to NMS | | 分子名称: | D(-)-TARTARIC ACID, Muscarinic acetylcholine receptor M3,Lysozyme,Muscarinic acetylcholine receptor M3, N-methyl scopolamine | | 著者 | Thorsen, T.S, Matt, R, Weis, W.I, Kobilka, B. | | 登録日 | 2014-07-15 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Modified T4 Lysozyme Fusion Proteins Facilitate G Protein-Coupled Receptor Crystallogenesis.
Structure, 22, 2014
|
|
4RYE
 
 | | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv | | 分子名称: | D-alanyl-D-alanine carboxypeptidase | | 著者 | Cuff, M, Tan, K, Hatzos-Skintges, C, Jedrzejczak, R, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2014-12-15 | | 公開日 | 2015-01-28 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | The crystal structure of D-ALANYL-D-ALANINE CARBOXYPEPTIDASE from Mycobacterium tuberculosis H37Rv
To be Published
|
|
4RYV
 
 | | Crystal structure of yellow lupin LLPR-10.1A protein in complex with trans-zeatin | | 分子名称: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Protein LLPR-10.1A, SULFATE ION | | 著者 | Dolot, R, Michalska, K, Sliwiak, J, Bujacz, G, Sikorski, M.M, Jaskolski, M. | | 登録日 | 2014-12-17 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Crystallographic and CD probing of ligand-induced conformational changes in a plant PR-10 protein.
J.Struct.Biol., 193, 2016
|
|
5LH6
 
 | | High dose Thaumatin - 360-400 ms. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
4TOH
 
 | | 1.80A resolution structure of Iron Bound BfrB (C89S, K96C) from Pseudomonas aeruginosa | | 分子名称: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | 登録日 | 2014-06-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
5LY1
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II) AND Macrocyclic PEPTIDE Inhibitor CP2 (13-mer) | | 分子名称: | CHLORIDE ION, CP2, GLYCEROL, ... | | 著者 | King, O.N.F, Chowdhury, R, Kawamura, A, Schofield, C.J. | | 登録日 | 2016-09-23 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
5DRM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a dichloro-substituted, 2,5-diarylthiophene-core ligand 4,4'-thiene-2,5-diylbis(3-chlorophenol) | | 分子名称: | 4,4'-thiene-2,5-diylbis(3-chlorophenol), Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-16 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.241 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5LI2
 
 | | bacteriophage phi812K1-420 tail sheath and tail tube protein in native tail | | 分子名称: | Phage-like element PBSX protein XkdM, tail sheath protein | | 著者 | Novacek, J, Siborova, M, Benesik, M, Pantucek, R, Doskar, J, Plevka, P. | | 登録日 | 2016-07-14 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5LJK
 
 | | Crystal structure of human apo CRBP1 | | 分子名称: | Retinol-binding protein 1, SODIUM ION | | 著者 | Zanotti, G, Vallese, F, Berni, R, Menozzi, I. | | 登録日 | 2016-07-18 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and molecular determinants affecting the interaction of retinol with human CRBP1.
J. Struct. Biol., 197, 2017
|
|
5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | 分子名称: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | 著者 | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | 登録日 | 2016-07-28 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
5LOP
 
 | | Structure of the active form of /K. lactis/ Dcp1-Dcp2-Edc3 decapping complex bound to m7GDP | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, KLLA0A11308p, KLLA0E01827p, ... | | 著者 | Charenton, C, Taverniti, V, Gaudon-Plesse, C, Back, R, Seraphin, B, Graille, M. | | 登録日 | 2016-08-09 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator.
Nat.Struct.Mol.Biol., 23, 2016
|
|
