2VL6
 
 | | STRUCTURAL ANALYSIS OF THE SULFOLOBUS SOLFATARICUS MCM PROTEIN N- TERMINAL DOMAIN | | Descriptor: | MINICHROMOSOME MAINTENANCE PROTEIN MCM, ZINC ION | | Authors: | Liu, W, Pucci, B, Rossi, M, Pisani, F.M, Ladenstein, R. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Sulfolobus Solfataricus Mcm Protein N-Terminal Domain.
Nucleic Acids Res., 36, 2008
|
|
7AA7
 
 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
2VBX
 
 | |
1AF6
 
 | | MALTOPORIN SUCROSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
1AI8
 
 | | HUMAN ALPHA-THROMBIN TERNARY COMPLEX WITH THE EXOSITE INHIBITOR HIRUGEN AND ACTIVE SITE INHIBITOR PHCH2OCO-D-DPA-PRO-BOROMPG | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUDIN IIIB, ... | | Authors: | Skordalakes, E, Dodson, G, Elgendy, S, Goodwin, C.A, Green, D, Tyrrel, R, Scully, M.F, Freyssinet, J, Kakkar, V.V, Deadman, J. | | Deposit date: | 1997-05-01 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships.
Protein Sci., 1, 1992
|
|
7VEL
 
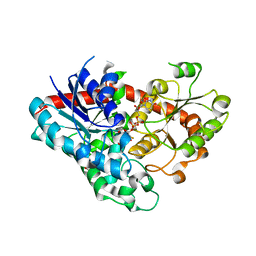 | | Crystal structure of Phytolacca americana UGT3 with UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VSQ
 
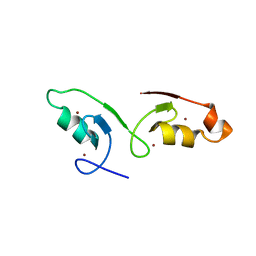 | |
6ZXS
 
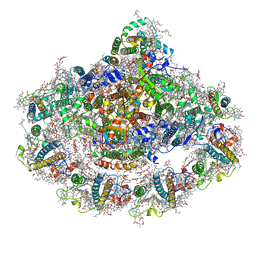 | | Cold grown Pea Photosystem I | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Borovikova-Sheinker, A, Subramanyam, R, Nelson, N. | | Deposit date: | 2020-07-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of cold grown pea Photosystem I
To Be Published
|
|
7VEK
 
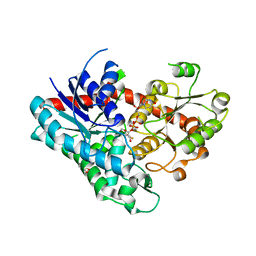 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VEJ
 
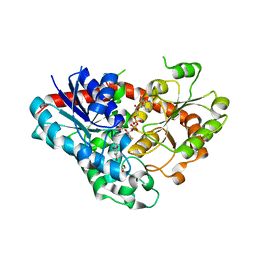 | | Crystal structure of Phytolacca americana UGT3 with kaempferol and UDP-2fluoroglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VD6
 
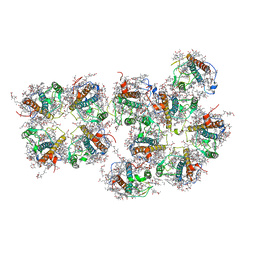 | | Structure of S1M1-type FCPII complex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
7VSP
 
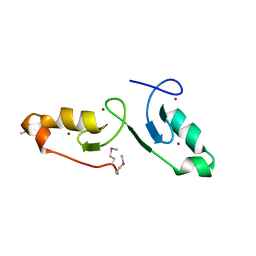 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
2VC1
 
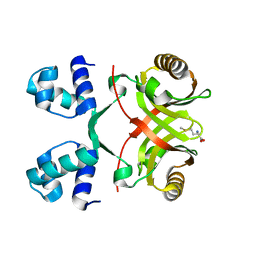 | |
2VBZ
 
 | |
7VU5
 
 | |
2VJI
 
 | | Tailspike protein of E.coli bacteriophage HK620 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CHLORIDE ION, ... | | Authors: | Mueller, J.J, Barbirz, S, Uetrecht, C, Seckler, R, Heinemann, U. | | Deposit date: | 2007-12-11 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of Escherichia Coli Phage Hk620 Tailspike: Podoviral Tailspike Endoglycosidase Modules are Evolutionarily Related.
Mol.Microbiol., 69, 2008
|
|
2V52
 
 | | Structure of MAL-RPEL2 complexed to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Guettler, S, Langer, C.A, Treisman, R, McDonald, N.Q. | | Deposit date: | 2008-10-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL.
EMBO J., 27, 2008
|
|
2VBY
 
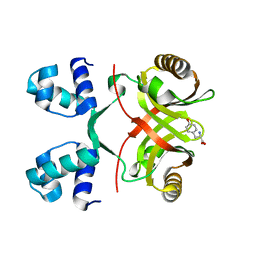 | |
3EOV
 
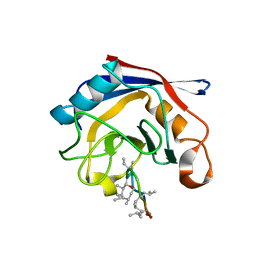 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6ZZ3
 
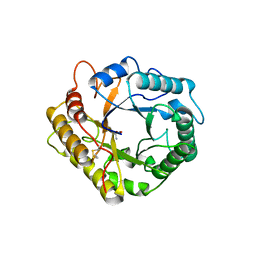 | | RBcel1 cellulase variant Y201F with cellotriose covalently bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Glycoside hydrolase family 5: structural snapshots highlighting the involvement of two conserved residues in catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VD5
 
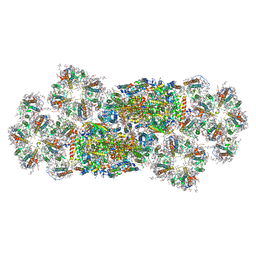 | | Structure of C2S2M2-type PSII-FCPII supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nagao, R, Kato, K, Akita, F, Miyazaki, N, Shen, J.R. | | Deposit date: | 2021-09-06 | | Release date: | 2022-03-02 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for different types of hetero-tetrameric light-harvesting complexes in a diatom PSII-FCPII supercomplex
Nat Commun, 13, 2022
|
|
2W84
 
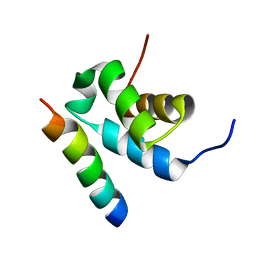 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
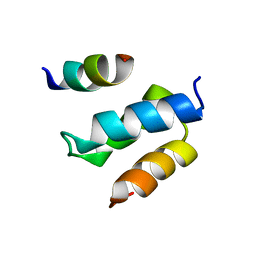 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6ZZY
 
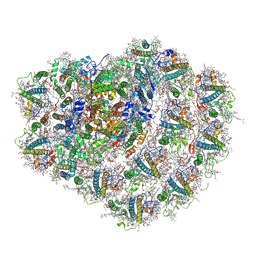 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
