7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
7JNR
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propionamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.443 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
4TOF
 
 | | 1.65A resolution structure of BfrB (C89S, K96C) crystal form 1 from Pseudomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterioferritin, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
7JNW
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | Descriptor: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.292 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JO2
 
 | | Carbonic Anhydrase IX Mimic Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | Descriptor: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Andring, J.T, McKenna, R. | | Deposit date: | 2020-08-05 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
5DIG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoromethyl-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-5-[4-hydroxy-2-(trifluoromethyl)phenyl]-7a-methyloctahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5ND8
 
 | | Hibernating ribosome from Staphylococcus aureus (Unrotated state) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khusainov, I, Vicens, Q, Ayupov, R, Usachev, K, Myasnikov, A, Simonetti, A, Validov, S, Kieffer, B, Yusupova, G, Yusupov, M, Hashem, Y. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
4TQE
 
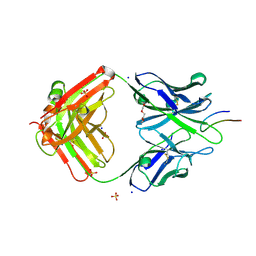 | |
5DJ2
 
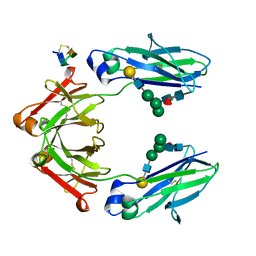 | | Fc Heterodimer Design 7.4 Y407A + T366V/K409V | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Secrist, E, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5DJD
 
 | | Fc Heterodimer Design 5.1 T366V + Y407F | | Descriptor: | Fc-III peptide, Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-02 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5D5A
 
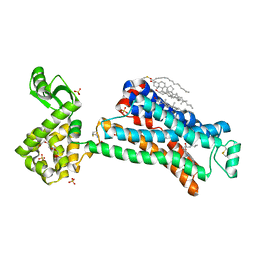 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5CV0
 
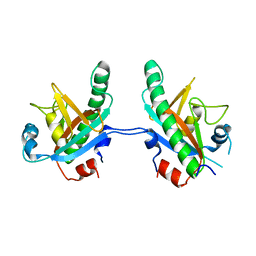 | | Crystal structure of N-terminal truncated human B12-chaperone CblD (108-296) | | Descriptor: | Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Yamada, K, Gherasim, C, Banerjee, R, Koutmos, M. | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human B12 Trafficking Protein CblD Reveals Molecular Mimicry and Identifies a New Subfamily of Nitro-FMN Reductases.
J.Biol.Chem., 290, 2015
|
|
7LMB
 
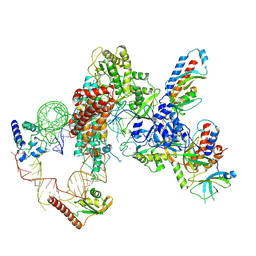 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
4TOB
 
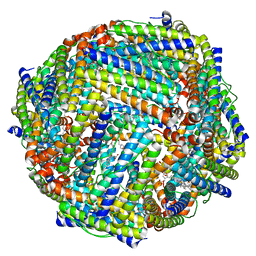 | | 1.95A resolution structure of BfrB (Q151L) from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TR5
 
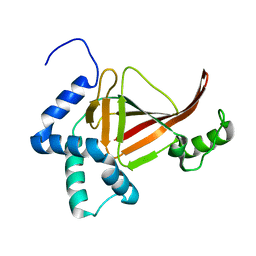 | |
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
4RWS
 
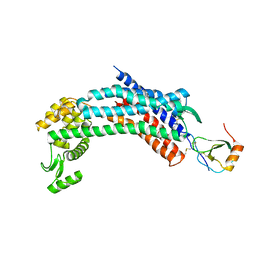 | | Crystal structure of CXCR4 and viral chemokine antagonist vMIP-II complex (PSI Community Target) | | Descriptor: | C-X-C chemokine receptor type 4/Endolysin chimeric protein, Viral macrophage inflammatory protein 2 | | Authors: | Qin, L, Kufareva, I, Holden, L, Wang, C, Zheng, Y, Wu, H, Fenalti, G, Han, G.W, Cherezov, V, Abagyan, R, Stevens, R.C, Handel, T.M, GPCR Network (GPCR) | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine.
Science, 347, 2015
|
|
5D3Q
 
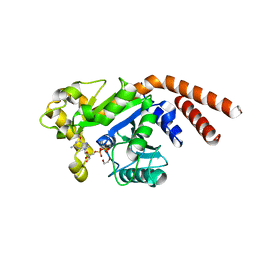 | | Dynamin 1 GTPase-BSE fusion dimer complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dynamin-1,Dynamin-1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Anand, R, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5NJ2
 
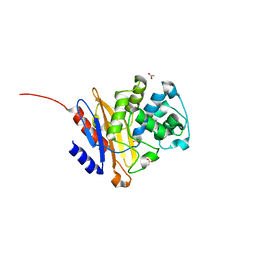 | | Crystal structure of BlaC from Mycobacterium tuberculosis bound to phosphate | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ... | | Authors: | Tassoni, R, Pannu, N.S, Ubbink, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Phosphate Promotes the Recovery of Mycobacterium tuberculosis beta-Lactamase from Clavulanic Acid Inhibition.
Biochemistry, 56, 2017
|
|
7LQ6
 
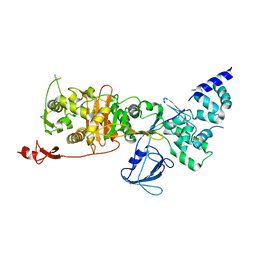 | | CryoEM structure of Escherichia coli PBP1b | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
4RZ0
 
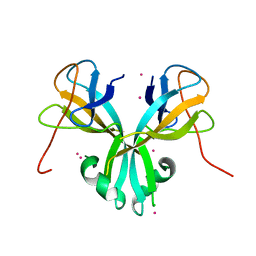 | | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c | | Descriptor: | PFL0690c, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Crystal Structure of Plasmodium falciparum putative histone methyltransferase PFL0690c
To be Published
|
|
5CUA
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(4-hydroxyphenyl)piperazin-1-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 1-Acetyl-4-(4-hydroxyphenyl)piperazine (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5NH0
 
 | |
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
4TKP
 
 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
