5HP6
 
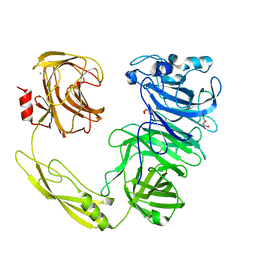 | | Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lansky, S, Salama, R, Shwartstien, O, Shoham, Y, Shoham, G. | | Deposit date: | 2016-01-20 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structure of AbnA, a GH43 extracellular arabinanase from Geobacillus stearothermophilus (a new conformational state)
To Be Published
|
|
5HQX
 
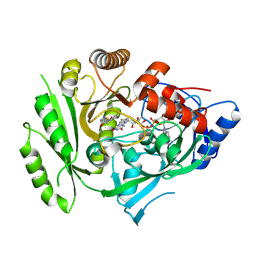 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor HETDZ | | Descriptor: | 1-[2-(2-hydroxyethyl)phenyl]-3-(1,2,3-thiadiazol-5-yl)urea, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P. | | Deposit date: | 2016-01-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase.
Plant Mol.Biol., 92, 2016
|
|
1EU8
 
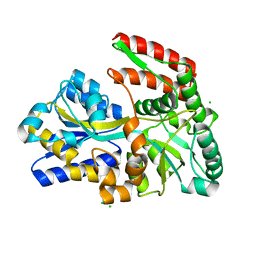 | | STRUCTURE OF TREHALOSE MALTOSE BINDING PROTEIN FROM THERMOCOCCUS LITORALIS | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, TREHALOSE/MALTOSE BINDING PROTEIN, ... | | Authors: | Diez, J, Diederichs, K, Greller, G, Horlacher, R, Boos, W, Welte, W. | | Deposit date: | 2000-04-14 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic Archaeon Thermococcus litoralis at 1.85 A.
J.Mol.Biol., 305, 2001
|
|
4XF6
 
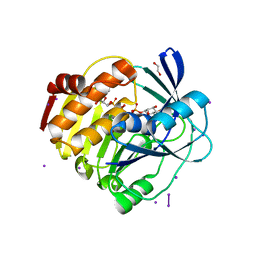 | | myo-inositol 3-kinase bound with its products (ADP and 1D-myo-inositol 3-phosphate) | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure and Product Analysis of an Archaeal myo-Inositol Kinase Reveal Substrate Recognition Mode and 3-OH Phosphorylation
Biochemistry, 54, 2015
|
|
8OTY
 
 | | Crystal structure of the titin domain Fn3-90 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
5HRT
 
 | | Crystal structure of mouse autotaxin in complex with a DNA aptamer | | Descriptor: | CALCIUM ION, CHLORIDE ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Kato, K, Nishimasu, H, Morita, J, Ishitani, R, Nureki, O. | | Deposit date: | 2016-01-24 | | Release date: | 2016-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis for specific inhibition of Autotaxin by a DNA aptamer
Nat.Struct.Mol.Biol., 23, 2016
|
|
8OT5
 
 | | Crystal structure of the titin domain Fn3-85 | | Descriptor: | CHLORIDE ION, SODIUM ION, Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8P0P
 
 | | Crystal structure of AaNGT complexed to UDP-2F-Glucose | | Descriptor: | Adhesin, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Piniello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8ORP
 
 | | Crystal structure of Drosophila melanogaster alpha-amylase in complex with the inhibitor acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of Drosophila melanogaster alpha-Amylase.
Molecules, 28, 2023
|
|
4XIX
 
 | | Carbonic anhydrase Cah3 from Chlamydomonas reinhardtii in complex with phosphate. | | Descriptor: | Carbonic anhydrase, alpha type, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hainzl, T, Grundstrom, C, Benlloch, R, Shevela, D, Shutova, T, Messinger, J, Samuelsson, G, Sauer-Eriksson, A.E. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas reinhardtii.
Plant Physiol., 167, 2015
|
|
1EYY
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
6ZBT
 
 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79948521 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
4XEQ
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (Deval_0042, TARGET EFI-510114) BOUND TO COPURIFIED (R)-PANTOIC ACID | | Descriptor: | PANTOATE, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-24 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO VULGARIS (Deval_0042, TARGET EFI-510114) BOUND TO COPURIFIED (R)-PANTOIC ACID
To be published
|
|
1EXB
 
 | | STRUCTURE OF THE CYTOPLASMIC BETA SUBUNIT-T1 ASSEMBLY OF VOLTAGE-DEPENDENT K CHANNELS | | Descriptor: | KV BETA2 PROTEIN, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM CHANNEL KV1.1 | | Authors: | Gulbis, J.M, Zhou, M, Mann, S, MacKinnon, R. | | Deposit date: | 2000-05-02 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the cytoplasmic beta subunit-T1 assembly of voltage-dependent K+ channels.
Science, 289, 2000
|
|
8OTM
 
 | | structure of InhA from mycobacterium tuberculosis in complex with N-((1-(3-hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-2-oxo-2H-chromene-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-~{N}-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methyl]chromene-3-carboxamide, ACETATE ION, ... | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
6ZAK
 
 | | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV. | | Descriptor: | Isopenicillin N synthase, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, SULFATE ION, ... | | Authors: | Rabe, P, Kamps, J.J.A.G, Sutherlin, K, Pharm, C, McDonough, M.A, Leissing, T.M, Aller, P, Butryn, A, Linyard, J, Lang, P, Brem, J, Fuller, F.D, Batyuk, A, Hunter, M.S, Pettinati, I, Clifton, I.J, Alonso-Mori, R, Gul, S, Young, I, Kim, I, Bhowmick, A, ORiordan, L, Brewster, A.S, Claridge, T.D.W, Sauter, N.K, Yachandra, V, Yano, J, Kern, J.F, Orville, A.M, Schofield, C.J. | | Deposit date: | 2020-06-05 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Room temperature XFEL Isopenicillin N synthase structure in complex with the Fe(IV)=O mimic VO and ACV.
To Be Published
|
|
1EWF
 
 | | THE 1.7 ANGSTROM CRYSTAL STRUCTURE OF BPI | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERICIDAL/PERMEABILITY-INCREASING PROTEIN | | Authors: | Kleiger, G, Beamer, L.J, Grothe, R, Mallick, P, Eisenberg, D. | | Deposit date: | 2000-04-25 | | Release date: | 2000-06-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of BPI: a study of how two dissimilar amino acid sequences can adopt the same fold.
J.Mol.Biol., 299, 2000
|
|
4XJX
 
 | | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HsdR, MAGNESIUM ION | | Authors: | Baikova, T, Stsiapanava, A, Moche, M, Degtjarik, O, Kuta-Smatanova, I, Ettrich, R. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP
To Be Published
|
|
2LDJ
 
 | | 1H Chemical Shift Assignments and structure of Trp-Cage mini-protein with D-amino acid | | Descriptor: | Trp-Cage mini-protein | | Authors: | Granillo, A.R, Annavarapu, S, Zhang, L, Koder, R, Nanda, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-11-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational Design of Thermostabilizing d-Amino Acid Substitutions.
J.Am.Chem.Soc., 133, 2011
|
|
3DCP
 
 | | Crystal structure of the putative histidinol phosphatase hisK from Listeria monocytogenes. Northeast Structural Genomics Consortium target LmR141. | | Descriptor: | FE (III) ION, Histidinol-phosphatase, ZINC ION | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Zhao, L, Mao, L, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-04 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the putative histidinol phosphatase hisK from Listeria monocytogenes.
To be Published
|
|
1ID8
 
 | | NMR STRUCTURE OF GLUTAMATE MUTASE (B12-BINDING SUBUNIT) COMPLEXED WITH THE VITAMIN B12 NUCLEOTIDE | | Descriptor: | 2-HYDROXY-PROPYL-AMMONIUM, METHYLASPARTATE MUTASE S CHAIN, PHOSPHORIC ACID MONO-[5-(5,6-DIMETHYL-BENZOIMIDAZOL-1-YL)-4-HYDROXY-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER | | Authors: | Tollinger, M, Eichmuller, C, Konrat, R, Huhta, M.S, Marsh, E.N.G, Krautler, B. | | Deposit date: | 2001-04-04 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B(12).
J.Mol.Biol., 309, 2001
|
|
4XO4
 
 | |
1MO1
 
 | |
2IV0
 
 | | Thermal stability of isocitrate dehydrogenase from Archaeoglobus fulgidus studied by crystal structure analysis and engineering of chimers | | Descriptor: | CHLORIDE ION, ISOCITRATE DEHYDROGENASE, ZINC ION | | Authors: | Stokke, R, Karlstrom, M, Yang, N, Leiros, I, Ladenstein, R, Birkeland, N.K, Steen, I.H. | | Deposit date: | 2006-06-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thermal Stability of Isocitrate Dehydrogenase from Archaeoglobus Fulgidus Studied by Crystal Structure Analysis and Engineering of Chimers
Extremophiles, 11, 2007
|
|
