8C9C
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L22, 50S ribosomal protein L24, ... | | Authors: | Nikolay, R, Qin, B, Lauer, S. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.62 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C91
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C8Z
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C92
 
 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C94
 
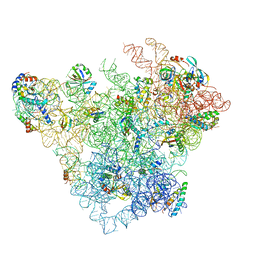 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C8Y
 
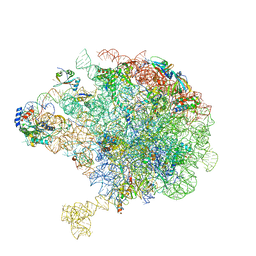 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C97
 
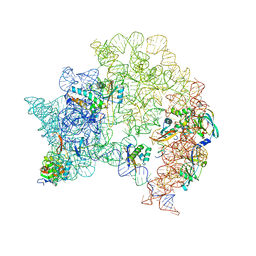 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C98
 
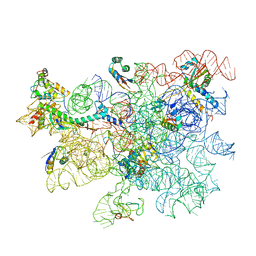 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C9A
 
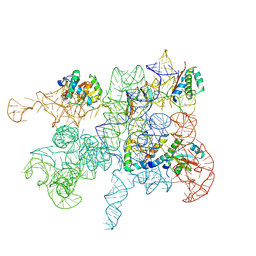 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L22, 50S ribosomal protein L23, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C90
 
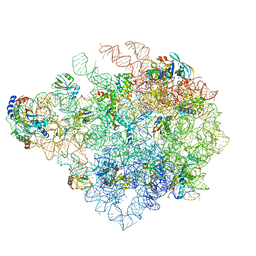 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C8X
 
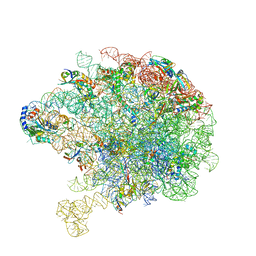 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
8C95
 
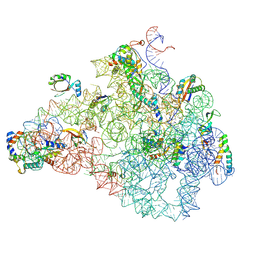 | | Cryo-EM captures early ribosome assembly in action | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Lauer, S, Nikolay, R, Qin, B. | | Deposit date: | 2023-01-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.92 Å) | | Cite: | Cryo-EM captures early ribosome assembly in action.
Nat Commun, 14, 2023
|
|
5XET
 
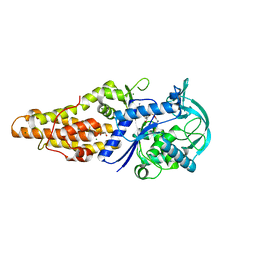 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
5Z3Q
 
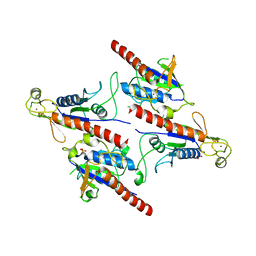 | | Crystal Structure of a Soluble Fragment of Poliovirus 2C ATPase (2.55 Angstrom) | | Descriptor: | PHOSPHATE ION, PV-2C, ZINC ION | | Authors: | Guan, H, Tian, J, Zhang, C, Qin, B, Cui, S. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Crystal structure of a soluble fragment of poliovirus 2CATPase
PLoS Pathog., 14, 2018
|
|
5Z0W
 
 | |
6J5E
 
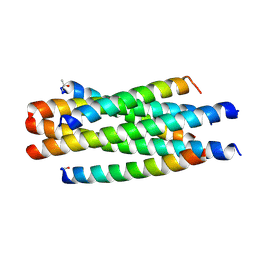 | |
6JQK
 
 | | Structure of C34M/N36 | | Descriptor: | C34M, N36 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-03-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of C34M/N36
To Be Published
|
|
6KTS
 
 | | Structure of C34N126K/N36 | | Descriptor: | Envelope glycoprotein, Glycoprotein 41 | | Authors: | Yu, D.W, Qin, B. | | Deposit date: | 2019-08-28 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Characterization of the Secondary Mutation N126K Selected by Various HIV-1 Fusion Inhibitors.
Viruses, 12, 2020
|
|
8HHK
 
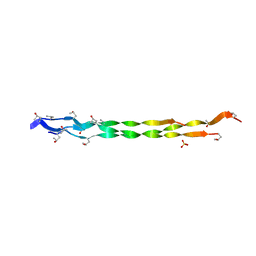 | |
8HHI
 
 | |
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | Descriptor: | ACETATE ION, Protein 2C | | Authors: | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2021-02-24 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
