5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3QZW
 
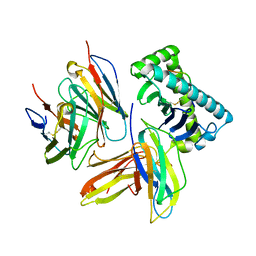 | | Plasticity of human CD8 binding to peptide-HLA-A*2402 | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-29 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Plasticity of human CD8alpha alpha binding to peptide-HLA-A*2402
Mol.Immunol., 48, 2011
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XOV
 
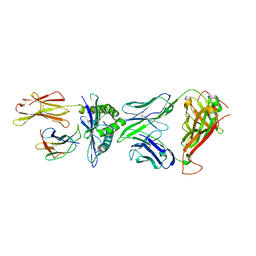 | | Crystal structure of peptide-HLA-A24 bound to S19-2 V-delta/V-beta TCR | | Descriptor: | Beta-2-microglobulin, HIV-1 Nef138-10 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.684 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XOT
 
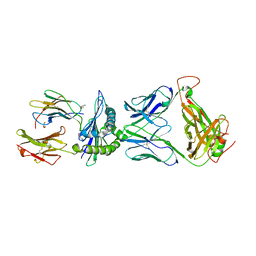 | | Crystal structure of pHLA-B35 in complex with TU55 T cell receptor | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5XOS
 
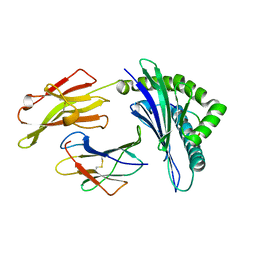 | | Crystal structure of HLA-B35 in complex with a pepetide antigen | | Descriptor: | An HIV reverse transcriptase epitope, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Conserved V delta 1 Binding Geometry in a Setting of Locus-Disparate pHLA Recognition by delta / alpha beta T Cell Receptors (TCRs): Insight into Recognition of HIV Peptides by TCRs.
J. Virol., 91, 2017
|
|
5GIX
 
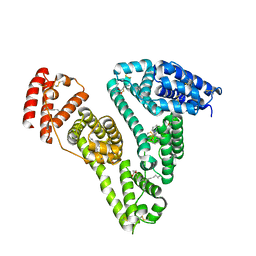 | | Human serum albumin-Palmitic acid-Fe(Hn3piT)Cl2 | | Descriptor: | 14-piperidin-1-yl-11-oxa-13$l^{3}-thia-15,16$l^{4}-diaza-12$l^{3}-ferratetracyclo[8.7.0.0^{2,7}.0^{12,16}]heptadeca-1(10),2(7),3,5,8,13,16-heptaene, PALMITIC ACID, Serum albumin | | Authors: | Yang, F, Qi, J, Wang, T. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Developing Anticancer Ferric Prodrugs Based on the N-Donor Residues of Human Serum Albumin Carrier IIA Subdomain
J. Med. Chem., 59, 2016
|
|
5GYQ
 
 | |
5H5N
 
 | |
4H53
 
 | | Influenza N2-Tyr406Asp neuraminidase in complex with beta-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
4H5S
 
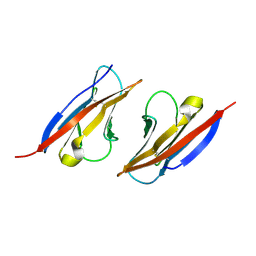 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
4H52
 
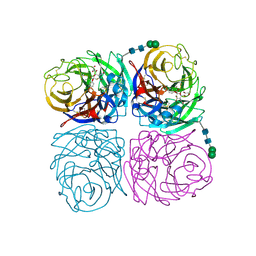 | | Wild-type influenza N2 neuraminidase covalent complex with 3-fluoro-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
4HX1
 
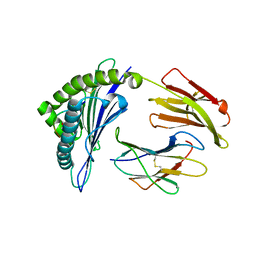 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4HWZ
 
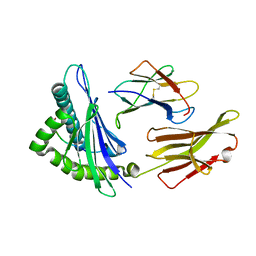 | | Structure of HLA-A68 complexed with an HIV derived peptide | | Descriptor: | 9-mer peptide from Pol protein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4I48
 
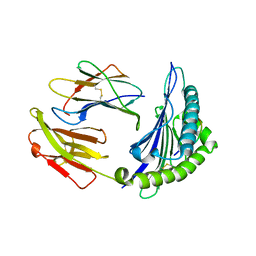 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4JTX
 
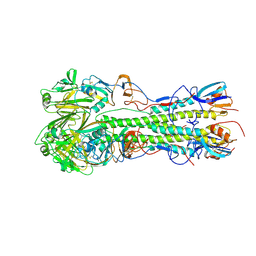 | | Crystal structure of 2009 pandemic influenza virus hemagglutinin mutant D225E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4JUJ
 
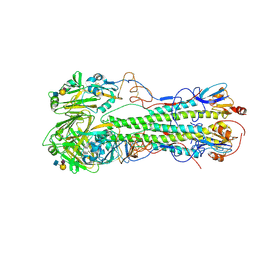 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4JUG
 
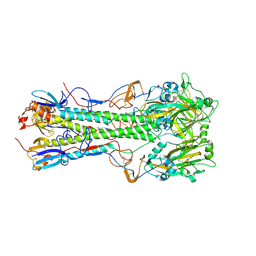 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4JU0
 
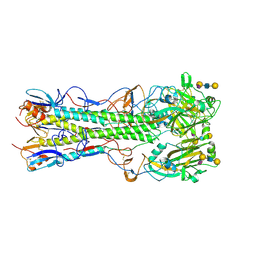 | | Crystal structure of 2009 pandemic influenza virus hemagglutinin mutant D225E complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4JUH
 
 | | Crystal structure of 1918 pandemic influenza virus hemagglutinin mutant D225G complexed with avian receptor analogue LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
4EWJ
 
 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
