5E5W
 
 | | Hemagglutinin-esterase-fusion mutant structure of influenza D virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
3UYW
 
 | | Crystal structures of globular head of 2009 pandemic H1N1 hemagglutinin | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Hemagglutinin | | Authors: | Xuan, C.L, Shi, Y, Qi, J.X, Xiao, H.X, Gao, G.F. | | Deposit date: | 2011-12-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines
Protein Cell, 2, 2011
|
|
4MWL
 
 | | Shanghai N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MX0
 
 | | Shanghai N9-peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
5E62
 
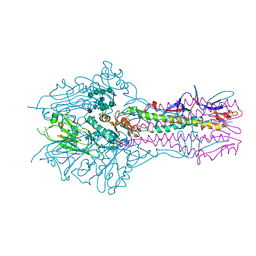 | | HEF-mut with Tr323 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-azidoethyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
5E66
 
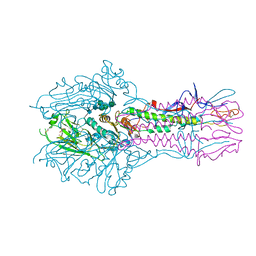 | | The complex structure of Hemagglutinin-esterase-fusion mutant protein from the influenza D virus with receptor analog 9-N-Ac-Sia | | Descriptor: | (6R)-5-acetamido-6-[(1S,2S)-3-acetamido-1,2-dihydroxypropyl]-3,5-dideoxy-beta-L-threo-hex-2-ulopyranosonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, H, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism
PLoS Pathog., 12, 2016
|
|
8X85
 
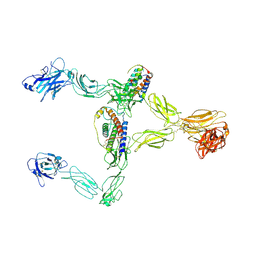 | | Structure of leptin-LepR dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, Leptin receptor, ... | | Authors: | Xie, Y.F, Shang, G.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
4MWY
 
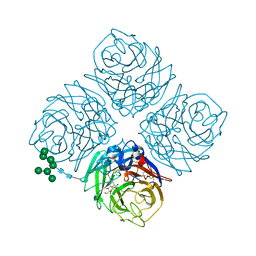 | | Shanghai N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4K1K
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
7XBY
 
 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
4K1H
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
4K1J
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
4O2C
 
 | | An Nt-acetylated peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
4QY0
 
 | | Structure of H10 from human-infecting H10N8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4QY2
 
 | | Structure of H10 from human-infecting H10N8 virus in complex with human receptor analog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid, hemagglutinin | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
3S91
 
 | | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain-containing protein 3, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD3 in complex with the inhibitor JQ1
To be Published
|
|
5GSX
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-2 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
8HIT
 
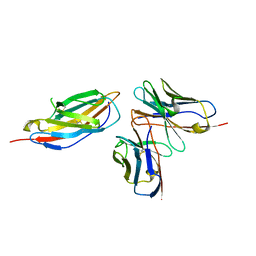 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | Authors: | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
5GSV
 
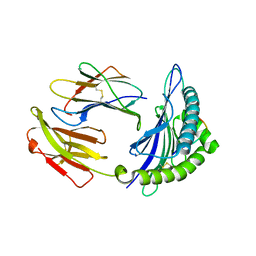 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | Descriptor: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GR7
 
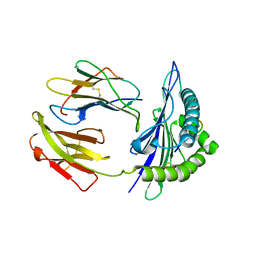 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSR
 
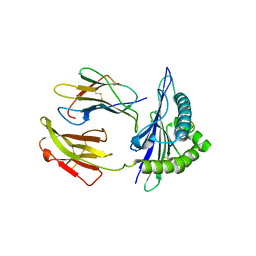 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | Descriptor: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-17 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GSB
 
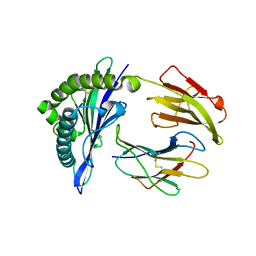 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-3 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | Authors: | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
