5CM8
 
 | |
5CM9
 
 | |
8BCJ
 
 | |
8BCI
 
 | |
1PBL
 
 | | STRUCTURE OF RIBONUCLEIC ACID, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*OMCP*OMGP*OMCP*OMGP*OMCP*OMG)-3') | | Authors: | Popenda, M, Biala, E, Milecki, J, Adamiak, R.W. | | Deposit date: | 1996-08-05 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2'-O-Me(CGCGCG)2 under low salt conditions.
Nucleic Acids Res., 25, 1997
|
|
6FRH
 
 | | Crystal structure of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | Replicative DNA helicase,Replicative DNA helicase | | Authors: | Popp, M.A, Blankenfeldt, W, Gazdag, M.E, Matern, J.J.C, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6FRE
 
 | | Crystal structure of G-1F/H73A mutant of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | Replicative DNA helicase,Replicative DNA helicase | | Authors: | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
6FRG
 
 | | Crystal structure of G-1F mutant of Ssp DnaB Mini-Intein variant M86 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Replicative DNA helicase, ... | | Authors: | Popp, M.A, Blankenfeldt, W, Friedel, K, Mootz, H.D. | | Deposit date: | 2018-02-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | A functional interplay between intein and extein sequences in protein splicing compensates for the essential block B histidine.
Chem Sci, 10, 2019
|
|
1PBM
 
 | | STRUCTURE OF RIBONUCLEIC ACID, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Popenda, M, Biala, E, Milecki, J, Adamiak, R.W. | | Deposit date: | 1996-08-05 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2'-O-Me(CGCGCG)2 under low salt conditions.
Nucleic Acids Res., 25, 1997
|
|
1T4X
 
 | | The first left-handed RNA structure of (CGCGCG)2, Z-RNA, NMR, 12 structures, determined in high salt | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Popenda, M, Milecki, J, Adamiak, R.W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High salt solution structure of a left-handed RNA double helix.
Nucleic Acids Res., 32, 2004
|
|
6TCB
 
 | |
8AJQ
 
 | |
8AID
 
 | |
2M1O
 
 | | ID3 stem | | Descriptor: | RNA (5'-R(P*AP*GP*CP*AP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*UP*GP*UP*A)-3') | | Authors: | Popovic, M, Greenbaum, N.L. | | Deposit date: | 2012-12-03 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ID3 stem
To be Published
|
|
2M12
 
 | |
6TC2
 
 | | Monoclinic human insulin in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | Authors: | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | Descriptor: | chemotaxis-inhibiting protein CHIPS | | Authors: | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
1UYJ
 
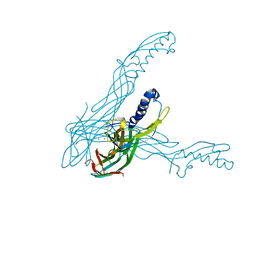 | | Clostridium perfringens epsilon toxin shows structural similarity with the pore forming toxin aerolysin | | Descriptor: | EPSILON-TOXIN, URANIUM ATOM | | Authors: | Cole, A.R, Gibert, M, Poppoff, M, Moss, D.S, Titball, R.W, Basak, A.K. | | Deposit date: | 2004-03-02 | | Release date: | 2004-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clostridium Perfringens Epsilon-Toxin Shows Structural Similarity to the Pore-Forming Toxin Aerolysin
Nat.Struct.Mol.Biol., 11, 2004
|
|
6RB9
 
 | | The pore structure of Clostridium perfringens epsilon toxin | | Descriptor: | Epsilon-toxin type B | | Authors: | Savva, C.G, Clark, A.R, Naylor, C.E, Popoff, M.R, Moss, D.S, Basak, A.K, Titball, R.W, Bokori-Brown, M. | | Deposit date: | 2019-04-09 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The pore structure of Clostridium perfringens epsilon toxin.
Nat Commun, 10, 2019
|
|
7ZSH
 
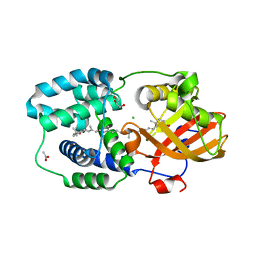 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 2 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSI
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 5 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSJ
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 10 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSF
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSG
 
 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 1 minute of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
2YGT
 
 | | Clostridium perfringens delta-toxin | | Descriptor: | DELTA TOXIN, GLYCEROL, IMIDAZOLE, ... | | Authors: | Huyet, J, Naylor, C.E, Gibert, M, Popoff, M.R, Basak, A.K. | | Deposit date: | 2011-04-20 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights Into Clostridium Perfringens Delta Toxin Pore Formation.
Plos One, 8, 2013
|
|
