8U49
 
 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
1IG8
 
 | | Crystal Structure of Yeast Hexokinase PII with the correct amino acid sequence | | Descriptor: | SULFATE ION, hexokinase PII | | Authors: | Kuser, P.R, Krauchenco, S, Antunes, O.A, Polikarpov, I. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action.
J.Biol.Chem., 275, 2000
|
|
1JV4
 
 | | Crystal structure of recombinant major mouse urinary protein (rmup) at 1.75 A resolution | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, Major urinary protein 2 | | Authors: | Kuser, P.R, Franzoni, L, Ferrari, E, Spisni, A, Polikarpov, I. | | Deposit date: | 2001-08-28 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray structure of a recombinant major urinary protein at 1.75 A resolution. A comparative study of X-ray and NMR-derived structures.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4XTA
 
 | |
7PXQ
 
 | | GH115 alpha-1,2-glucuronidase D303A | | Descriptor: | CALCIUM ION, xylan alpha-1,2-glucuronidase | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4TUF
 
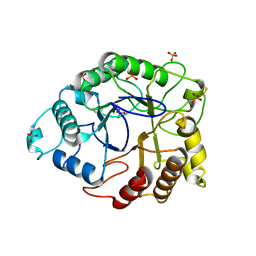 | |
8U4A
 
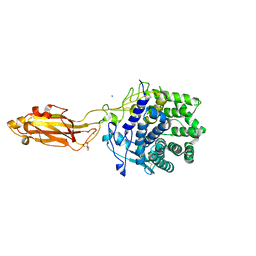 | |
6MVI
 
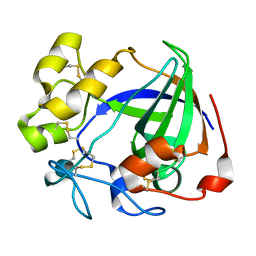 | | Apo Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
6MVJ
 
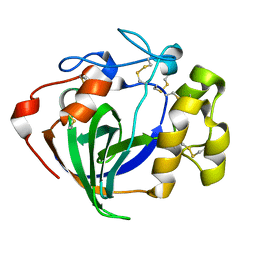 | | Cellobiose complex Cel45A from Neurospora crassa OR74A | | Descriptor: | Endoglucanase V, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2018-10-25 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Structural insights into the hydrolysis pattern and molecular dynamics simulations of GH45 subfamily a endoglucanase from Neurospora crassa OR74A.
Biochimie, 165, 2019
|
|
3NEE
 
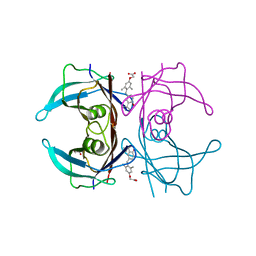 | | Wild type human transthyretin (TTR) complexed with GC-1 (TTRwt:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
2RC5
 
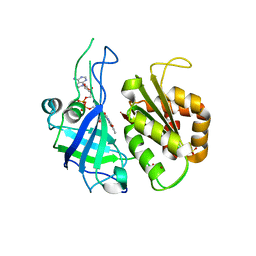 | | Refined structure of FNR from Leptospira interrogans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, SULFATE ION, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Polikarpov, I, Ceccarelli, E.A. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
2RC6
 
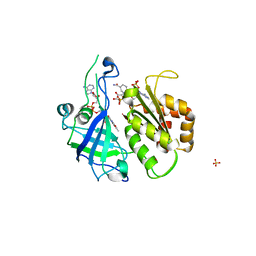 | | Refined structure of FNR from Leptospira interrogans bound to NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Ceccarelli, E.A, Polikarpov, I. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
8FAA
 
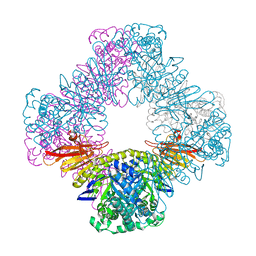 | |
8FA5
 
 | |
4UCF
 
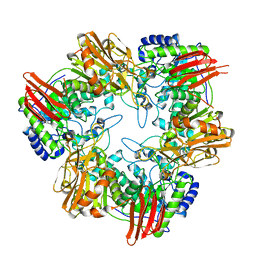 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
4UOJ
 
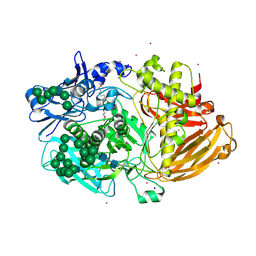 | | Structure of Fungal beta-mannosidase (GH2) from Trichoderma harzianum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(3-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Muniz, J.R.C, Aparicio, R, Santos, J.C, Nascimento, A.S, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into the Structure and Function of Fungal Beta-Mannosidases from Glycoside Hydrolase Family 2 Based on Multiple Crystal Structures of the Trichoderma Harzianum Enzyme.
FEBS J., 281, 2014
|
|
1L9W
 
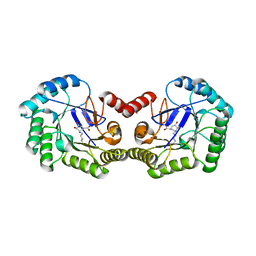 | | CRYSTAL STRUCTURE OF 3-DEHYDROQUINASE FROM SALMONELLA TYPHI COMPLEXED WITH REACTION PRODUCT | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase aroD | | Authors: | Lee, W.H, Perles, L.A, Nagem, R.A.P, Shrive, A.K, Hawkins, A, Sawyer, L, Polikarpov, I. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
8FA4
 
 | |
1Y9M
 
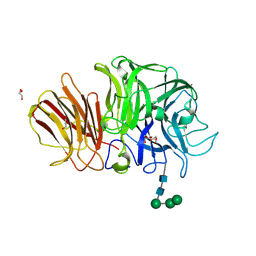 | | Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Nagem, R.A.P, Rojas, A.L, Golubev, A.M, Korneeva, O.S, Eneyskaya, E.V, Kulminskaya, A.A, Neustroev, K.N, Polikarpov, I. | | Deposit date: | 2004-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of exo-inulinase from Aspergillus awamori: the enzyme fold and structural determinants of substrate recognition
J.Mol.Biol., 344, 2004
|
|
1XC6
 
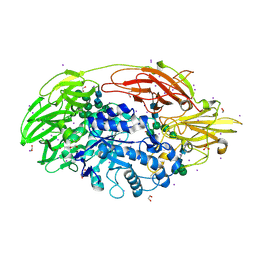 | | Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rojas, A.L, Nagem, R.A.P, Neustroev, K.N, Arand, M, Adamska, M, Eneyskaya, E.V, Kulminskaya, A.A, Garratt, R.C, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of beta-Galactosidase from Penicillium sp. and its Complex with Galactose
J.Mol.Biol., 343, 2004
|
|
3SZ1
 
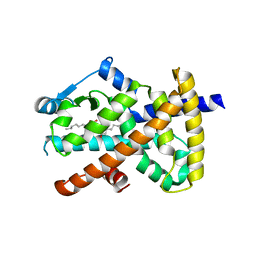 | | Human PPAR gamma ligand binding domain in complex with luteolin and myristic acid | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, MYRISTIC ACID, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Puhl, A.C, Bernardes, A, Polikarpov, I. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mode of peroxisome proliferator-activated receptor gamma activation by luteolin.
Mol.Pharmacol., 81, 2012
|
|
3TKM
 
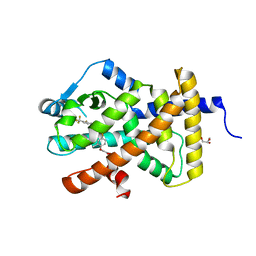 | | Crystal structure PPAR delta binding GW0742 | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor delta, {4-[({2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-1,3-thiazol-5-yl}methyl)sulfanyl]-2-methylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Batista, F.H, Polikarpov, I. | | Deposit date: | 2011-08-27 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural Insights into Human Peroxisome Proliferator Activated Receptor Delta (PPAR-Delta) Selective Ligand Binding.
Plos One, 7, 2012
|
|
2P3D
 
 | | Crystal Structure of the multi-drug resistant mutant subtype F HIV protease complexed with TL-3 inhibitor | | Descriptor: | Pol protein, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P3A
 
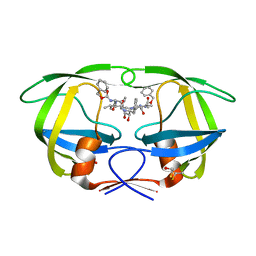 | | Crystal Structure of the multi-drug resistant mutant subtype B HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
2P3B
 
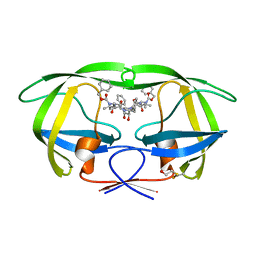 | | Crystal Structure of the subtype B wild type HIV protease complexed with TL-3 inhibitor | | Descriptor: | benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate, protease | | Authors: | Sanches, M, Krauchenco, S, Martins, N.H, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of B and non-B Subtypes of HIV-Protease: Insights into the Natural Susceptibility to Drug Resistance Development.
J.Mol.Biol., 369, 2007
|
|
