1QY2
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOPROPYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1QY1
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1CMA
 
 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
1GOH
 
 | |
1GOG
 
 | |
1GOF
 
 | | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | | Descriptor: | ACETIC ACID, COPPER (II) ION, GALACTOSE OXIDASE, ... | | Authors: | Ito, N, Phillips, S.E.V, Knowles, P.F. | | Deposit date: | 1993-09-30 | | Release date: | 1994-01-31 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase.
Nature, 350, 1991
|
|
1F9N
 
 | | CRYSTAL STRUCTURE OF AHRC, THE ARGININE REPRESSOR/ACTIVATOR PROTEIN FROM BACILLUS SUBTILIS | | Descriptor: | ARGININE REPRESSOR/ACTIVATOR PROTEIN | | Authors: | Dennis, C.A, Glykos, N.M, Parsons, M.R, Phillips, S.E.V. | | Deposit date: | 2000-07-11 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1CFV
 
 | | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | | Descriptor: | ESTRONE BETA-D-GLUCURONIDE, MONOCLONAL ANTIBODY FV4155, ZINC ION | | Authors: | Trinh, C.H, Phillips, S.E.V. | | Deposit date: | 1997-04-11 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity.
Structure, 5, 1997
|
|
1EFC
 
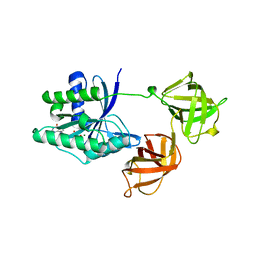 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1MUP
 
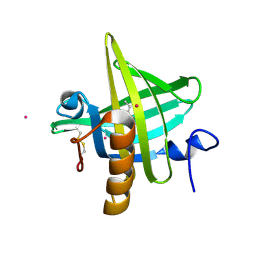 | | PHEROMONE BINDING TO TWO RODENT URINARY PROTEINS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN | | Authors: | Bocskei, Z, Flower, D.R, Groom, C.R, Phillips, S.E.V, North, A.C.T. | | Deposit date: | 1992-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography.
Nature, 360, 1992
|
|
1DMU
 
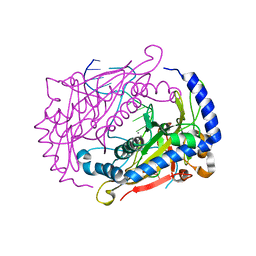 | | Crystal structure of the restriction endonuclease BglI (e.c.3.1.21.4) bound to its dna recognition sequence | | Descriptor: | BETA-MERCAPTOETHANOL, BGLI RESTRICTION ENDONUCLEASE, CALCIUM ION, ... | | Authors: | Newman, M, Lunnen, K, Wilson, G, Greci, J, Schildkraut, I, Phillips, S.E.V. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-18 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence.
EMBO J., 17, 1998
|
|
1QAL
 
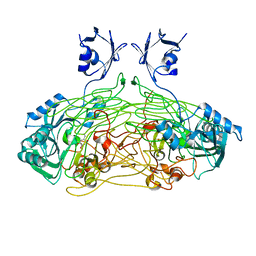 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-19 | | Release date: | 1999-08-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1FZR
 
 | | CRYSTAL STRUCTURE OF BACTERIOPHAGE T7 ENDONUCLEASE I | | Descriptor: | ENDONUCLEASE I | | Authors: | Hadden, J.M, Convery, M.A, Declais, A.C, Lilley, D.M.J, Phillips, S.E.V. | | Deposit date: | 2000-10-04 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I.
Nat.Struct.Biol., 8, 2001
|
|
1QAF
 
 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-11 | | Release date: | 1999-08-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1CMC
 
 | |
1YP7
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1ZSE
 
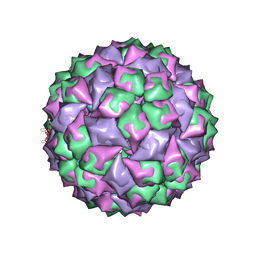 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
1OAC
 
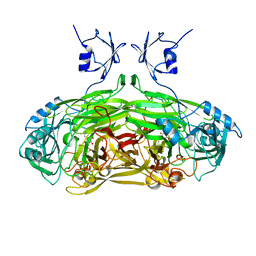 | | CRYSTAL STRUCTURE OF A QUINOENZYME: COPPER AMINE OXIDASE OF ESCHERICHIA COLI AT 2 ANGSTROEMS RESOLUTION | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Parsons, M.R, Convery, M.A, Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1995-09-27 | | Release date: | 1996-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution.
Structure, 3, 1995
|
|
1MJ2
 
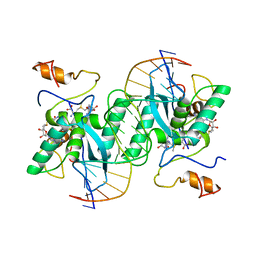 | |
1MJO
 
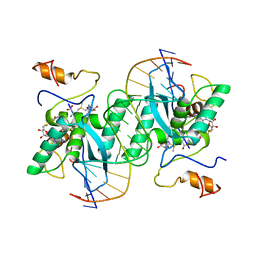 | |
1PKL
 
 | | THE STRUCTURE OF LEISHMANIA PYRUVATE KINASE | | Descriptor: | PROTEIN (PYRUVATE KINASE), SULFATE ION | | Authors: | Rigden, D.J, Phillips, S.E.V, Michels, P.A.M, Fothergill-Gilmore, L.A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of pyruvate kinase from Leishmania mexicana reveals details of the allosteric transition and unusual effector specificity.
J.Mol.Biol., 291, 1999
|
|
1DYU
 
 | | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2000-02-08 | | Release date: | 2000-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Active Site Base Controls Cofactor Reactivity in Escherichia Coli Amine Oxidase : X-Ray Crystallographicstudies with Mutational Variants
Biochemistry, 38, 1999
|
|
2JKX
 
 | | Galactose oxidase. MatGO. Copper free, expressed in Pichia Pastoris. | | Descriptor: | ACETATE ION, CALCIUM ION, GALACTOSE OXIDASE | | Authors: | Deacon, S.E, Mahmoud, K, Spooner, R.K, Firbank, S.J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Enhanced Fructose Oxidase Activity in a Galactose Oxidase Variant
Chembiochem, 5, 2004
|
|
