6XNC
 
 | |
8D5D
 
 | | Structure of Y430F D-ornithine/D-lysine decarboxylase complex with D-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-D-arginine, D-ornithine/D-lysine decarboxylase, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S, Nguyen Hoang, K.N. | | Deposit date: | 2022-06-04 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Y430F mutant of Salmonella d-ornithine/d-lysine decarboxylase has altered stereospecificity and a putrescine allosteric activation site.
Arch.Biochem.Biophys., 731, 2022
|
|
5JV3
 
 | | The neck-linker and alpha 7 helix of Homo sapiens Eg5 fused to EB1 | | Descriptor: | CALCIUM ION, Chimera protein of Kinesin-like protein KIF11 and Microtubule-associated protein RP/EB family member 1 | | Authors: | Phillips, R.K, Rayment, I. | | Deposit date: | 2016-05-10 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil.
J.Biol.Chem., 291, 2016
|
|
5JVS
 
 | |
5JVU
 
 | |
5JVR
 
 | |
5JVM
 
 | | The neck-linker and alpha 7 helix of Mus musculus KIF3C | | Descriptor: | Chimera protein of Kinesin-like protein KIF3C and Microtubule-associated protein RP/EB family member 1, MAGNESIUM ION | | Authors: | Phillips, R.K, Rayment, I. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.567 Å) | | Cite: | Family-specific Kinesin Structures Reveal Neck-linker Length Based on Initiation of the Coiled-coil.
J.Biol.Chem., 291, 2016
|
|
6XOY
 
 | |
7LC0
 
 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | Descriptor: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2021-01-09 | | Release date: | 2021-06-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
7LCE
 
 | | Structure of D-Glucosaminate-6-phosphate Ammonia-lyase | | Descriptor: | CALCIUM ION, D-glucosaminate-6-phosphate ammonia lyase, DIMETHYL SULFOXIDE | | Authors: | Phillips, R.S. | | Deposit date: | 2021-01-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.
Biochemistry, 60, 2021
|
|
6DVX
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant complexed with L-phenylalanine | | Descriptor: | (2E)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}-3-phenylpropanoic acid, (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-phenylalanine, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6DUR
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with L-phenylalanine | | Descriptor: | (2E)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}-3-phenylpropanoic acid, (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-phenylalanine, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-06-21 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6DXV
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant | | Descriptor: | POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Phillips, R.S. | | Deposit date: | 2018-07-01 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6DYT
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant complexed with L-alanine | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ALANYL-PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-07-02 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6DZ5
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant complexed with L-alanine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6ECG
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant complexed with L-methionine | | Descriptor: | (2E)-2-{[(Z)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4(1H)-YLIDENE}METHYL]IMINO}-4-(METHYLSULFANYL)BUTANOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
3E9K
 
 | | Crystal structure of Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex | | Descriptor: | 3-Hydroxyhippuric acid, Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Kumar, S, Gawandi, V, Momany, C, Phillips, R.S. | | Deposit date: | 2008-08-22 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity.
J.Med.Chem., 52, 2009
|
|
2HZP
 
 | | Crystal Structure of Homo Sapiens Kynureninase | | Descriptor: | Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Homo sapiens Kynureninase.
Biochemistry, 46, 2007
|
|
7UTC
 
 | |
7UUT
 
 | | Ternary complex crystal structure of secondary alcohol dehydrogenases from the Thermoanaerobacter ethanolicus mutants C295A and I86A provides better understanding of catalytic mechanism | | Descriptor: | (2R)-pentan-2-ol, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Dinh, T, Phillips, R, Rahn, K. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallographic snapshots of ternary complexes of thermophilic secondary alcohol dehydrogenase from Thermoanaerobacter pseudoethanolicus reveal the dynamics of ligand exchange and the proton relay network.
Proteins, 90, 2022
|
|
6XNB
 
 | |
3FDD
 
 | | The Crystal Structure of the Pseudomonas dacunhae Aspartate-Beta-Decarboxylase Reveals a Novel Oligomeric Assembly for a Pyridoxal-5-Phosphate Dependent Enzyme | | Descriptor: | ACETATE ION, CHLORIDE ION, L-aspartate-beta-decarboxylase, ... | | Authors: | Lima, S, Sundararaju, B, Huang, C, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the Pseudomonas dacunhae aspartate-beta-decarboxylase dodecamer reveals an unknown oligomeric assembly for a pyridoxal-5'-phosphate-dependent enzyme.
J.Mol.Biol., 388, 2009
|
|
1QZ9
 
 | | The Three Dimensional Structure of Kynureninase from Pseudomonas fluorescens | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, KYNURENINASE, ... | | Authors: | Momany, C, Levdikov, V, Blagova, L, Lima, S, Phillips, R.S. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of Kynureninase from Pseudomonas fluorescens.
Biochemistry, 43, 2004
|
|
4NOD
 
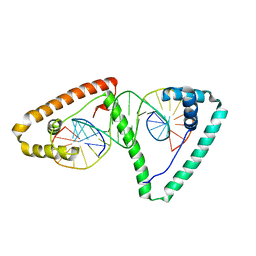 | | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation | | Descriptor: | 5'-D(*CP*CP*AP*AP*CP*CP*AP*AP*GP*CP*CP*CP*CP*AP*TP*AP*CP*CP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*GP*GP*TP*AP*TP*GP*GP*GP*GP*CP*TP*TP*GP*GP*(BRU)P*TP*GP*G)-3', Transcription factor A, ... | | Authors: | Ngo, H.B, Lovely, G.A, Phillips, R, Chan, D.C. | | Deposit date: | 2013-11-19 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation.
Nat Commun, 5, 2014
|
|
2YCP
 
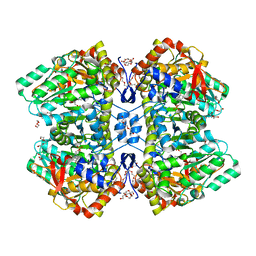 | | F448H mutant of tyrosine phenol-lyase from Citrobacter freundii in complex with quinonoid intermediate formed with 3-fluoro-L-tyrosine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Milic, D, Demidkina, T.V, Faleev, N.G, Phillips, R.S, Matkovic-Calogovic, D, Antson, A.A. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Snapshots of Tyrosine Phenol-Lyase Show that Substrate Strain Plays a Role in C-C Bond Cleavage
J.Am.Chem.Soc., 133, 2011
|
|
