4MGR
 
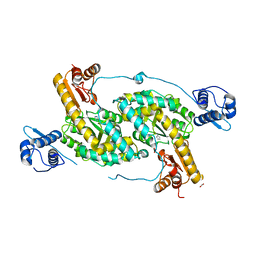 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J5F
 
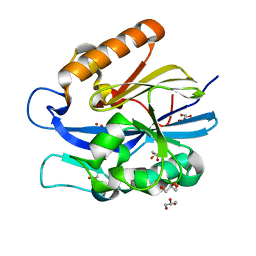 | | Crystal Structure of B. thuringiensis AiiA mutant F107W | | Descriptor: | GLYCEROL, N-acyl homoserine lactonase, ZINC ION | | Authors: | Liu, C.F, Liu, D, Momb, J, Thomas, P.W, Lajoie, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2013-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-gamma-lactonase from Bacillus thuringiensis.
Biochemistry, 52, 2013
|
|
4QYX
 
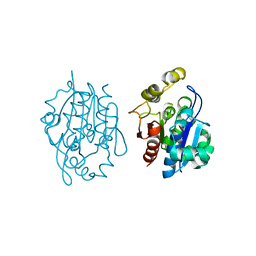 | | Crystal structure of YDR533Cp | | Descriptor: | Probable chaperone protein HSP31 | | Authors: | Wilson, M.A, Amour, S.T, Collins, J.L, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4PTH
 
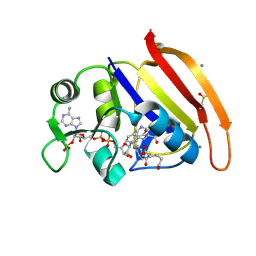 | | Ensemble model for Escherichia coli dihydrofolate reductase at 100K | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Keedy, D.A, van den Bedem, H, Sivak, D.A, Petsko, G.A, Ringe, D, Wilson, M.A, Fraser, J.S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Crystal Cryocooling Distorts Conformational Heterogeneity in a Model Michaelis Complex of DHFR.
Structure, 22, 2014
|
|
2AI0
 
 | | Anti-Cocaine Antibody 7.5.21, Crystal Form III | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain kappa, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-28 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
4RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
4RAT
 
 | |
4N0B
 
 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JKJ
 
 | |
2DAB
 
 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | D-AMINO ACID AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
1LRT
 
 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF TRITRICHOMONAS FOETUS INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE: STRUCTURAL CHARACTERIZATION OF NAD+ SITE IN MICROBIAL ENZYME | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, ... | | Authors: | Gan, L, Petsko, G.A, Hedstrom, L. | | Deposit date: | 2002-05-15 | | Release date: | 2003-07-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a ternary complex of Tritrichomonas foetus inosine
5'-monophosphate dehydrogenase: NAD+ orients the active site loop for catalysis
Biochemistry, 41, 2003
|
|
2PRQ
 
 | | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, COBALT (II) ION | | Authors: | Munih, P, Moulin, A, Stamper, C.C, Bennet, B, Ringe, D, Petsko, G.A, Holz, R.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica.
J.Inorg.Biochem., 101, 2007
|
|
1MUW
 
 | |
1LOK
 
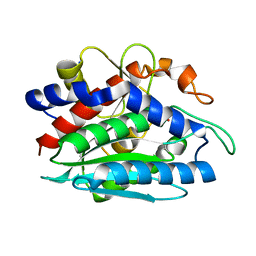 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | Authors: | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
1SBC
 
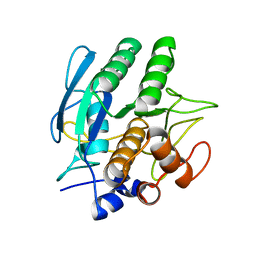 | |
2RAT
 
 | |
1OT5
 
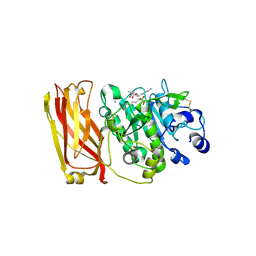 | | The 2.4 Angstrom Crystal Structure of Kex2 in complex with a peptidyl-boronic acid inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ac-Ala-Lys-boroArg N-acetylated boronic acid peptide inhibitor, ... | | Authors: | Holyoak, T, Wilson, M.A, Fenn, T.D, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-06-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A Resolution Crystal Structure of the Prototypical Hormone-Processing Protease Kex2 in Complex with an Ala-Lys-Arg Boronic Acid Inhibitor
Biochemistry, 42, 2003
|
|
2A1W
 
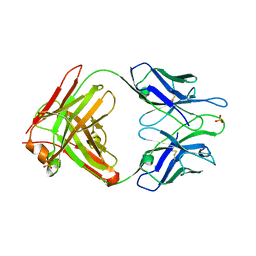 | | Anti-cocaine antibody 7.5.21, crystal form I | | Descriptor: | SULFATE ION, immunoglobulin heavy chain, immunoglobulin light chain kappa | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-06-21 | | Release date: | 2005-06-28 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexibility Of Packing:
Four Crystal Forms Of An Anti-Cocaine Antibody 7.5.21
To be Published
|
|
2A77
 
 | | Anti-Cocaine Antibody 7.5.21, Crystal Form II | | Descriptor: | GLYCEROL, Immunoglobulin Heavy Chain, Immunoglobulin Light Chain, ... | | Authors: | Pozharski, E, Hewagama, A, Shanafelt, A, Ringe, D, Petsko, G.A. | | Deposit date: | 2005-07-04 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flexibility of Packing: Four Crystal Forms of an Anti-Cocaine Antibody 7.5.21
To be Published
|
|
2A7M
 
 | | 1.6 Angstrom Resolution Structure of the Quorum-Quenching N-Acyl Homoserine Lactone Hydrolase of Bacillus thuringiensis | | Descriptor: | GLYCEROL, N-acyl homoserine lactone hydrolase, ZINC ION | | Authors: | Liu, D, Lepore, B.W, Petsko, G.A, Thomas, P.W, Stone, E.M, Fast, W, Ringe, D. | | Deposit date: | 2005-07-05 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2FOH
 
 | | Structure of porcine pancreatic elastase in 40% trifluoroethanol | | Descriptor: | CALCIUM ION, SULFATE ION, TRIFLUOROETHANOL, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
1P5F
 
 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Wilson, M.A, Collins, J.L, Hod, Y, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-04-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RKX
 
 | | Crystal Structure at 1.8 Angstrom of CDP-D-glucose 4,6-dehydratase from Yersinia pseudotuberculosis | | Descriptor: | CDP-glucose-4,6-dehydratase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Vogan, E.M, Bellamacina, C, He, X, Liu, H.W, Ringe, D, Petsko, G.A. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure at 1.8 A Resolution of CDP-d-Glucose 4,6-Dehydratase from Yersinia pseudotuberculosis
Biochemistry, 43, 2004
|
|
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
1SOA
 
 | | Human DJ-1 with sulfinic acid | | Descriptor: | RNA-binding protein regulatory subunit; oncogene DJ1 | | Authors: | Canet-Aviles, R, Wilson, M.A, Miller, D.W, Ahmad, R, McLendon, C, Bandyopadhyay, S, Baptista, M.J, Ringe, D, Petsko, G.A, Cookson, M.R. | | Deposit date: | 2004-03-13 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
