7T1J
 
 | | Crystal structure of RUBISCO from Rhodospirillaceae bacterium BRH_c57 | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Pereira, J.H, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural plasticity enables evolution and innovation of RuBisCO assemblies.
Sci Adv, 8, 2022
|
|
7T1C
 
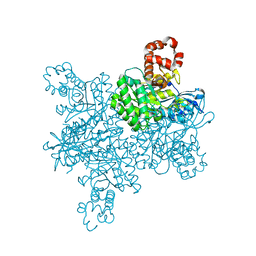 | |
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polyketide synthase-based biosynthesis of diols and aldehyde derivatives via terminal thioreduction
To Be Published
|
|
4NBV
 
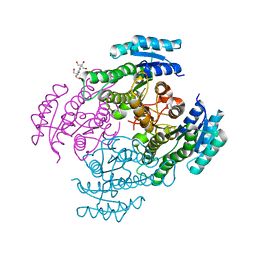 | | Crystal structure of FabG from Cupriavidus taiwanensis | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-[acyl-carrier-protein] reductase putative short-chain dehydrogenases/reductases (SDR) family protein | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBW
 
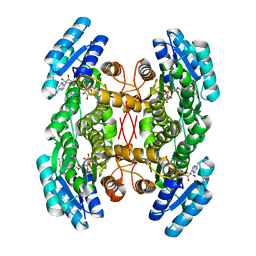 | | Crystal structure of FabG from Plesiocystis pacifica | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBU
 
 | | Crystal structure of FabG from Bacillus sp | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase, ACETOACETYL-COENZYME A | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBT
 
 | | Crystal structure of FabG from Acholeplasma laidlawii | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
7UOR
 
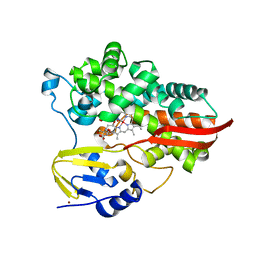 | | Crystal structure of cytochrome P450 enzyme CYP119 in complex with methyliridium(III) mesoporphyrin. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome, NICKEL (II) ION, ... | | Authors: | Pereira, J.H, Bloomer, B.J, Hartwig, J.F, Adams, P.D. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Mechanistic and structural characterization of an iridium-containing cytochrome reveals kinetically relevant cofactor dynamics
Nat Catal, 6, 2023
|
|
9BCM
 
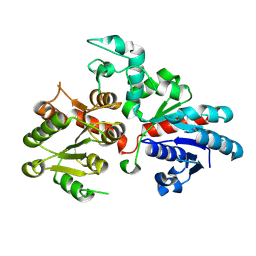 | |
4K41
 
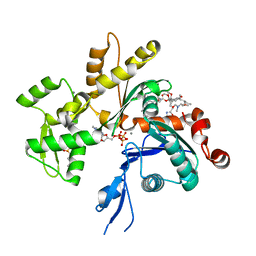 | | Crystal structure of actin in complex with marine macrolide kabiramide C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Pereira, J.H, Petchprayoon, C, Moriarty, N.W, Fink, S.J, Cecere, G, Paterson, I, Adams, P.D, Marriott, G. | | Deposit date: | 2013-04-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical studies of actin in complex with synthetic macrolide tail analogues.
Chemmedchem, 9, 2014
|
|
9B99
 
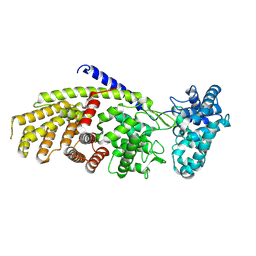 | | Crystal structure of Grindelia robusta 7,13-copalyl diphosphate synthase | | Descriptor: | Labda-7,13E-dienyl diphosphate synthase | | Authors: | Pereira, J.H, Cowie, A.E, Zerbe, P, Adams, P.D. | | Deposit date: | 2024-04-01 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Grindelia robusta 7,13-copalyl diphosphate synthase reveals active site features controlling catalytic specificity.
J.Biol.Chem., 300, 2024
|
|
4K42
 
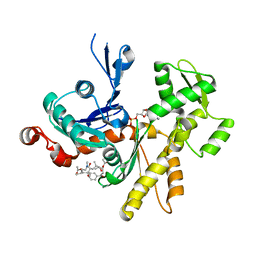 | | Crystal structure of actin in complex with synthetic AplC tail analogue SF01 [(3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate] | | Descriptor: | (3R,4S,5R,6S,10R,11R,12R)-11-(acetyloxy)-1-(benzyloxy)-14-[formyl(methyl)amino]-5-hydroxy-4,6,10,12-tetramethyl-9-oxotetradecan-3-yl propanoate, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Pereira, J.H, Petchprayoon, C, Moriarty, N.W, Fink, S.J, Cecere, G, Paterson, I, Adams, P.D, Marriott, G. | | Deposit date: | 2013-04-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical studies of actin in complex with synthetic macrolide tail analogues.
Chemmedchem, 9, 2014
|
|
4K43
 
 | | Crystal structure of actin in complex with synthetic AplC tail analogue GC04 [N-{(1E,4R,5R,7E,9S,10S,11S)-4,10-dimethoxy-11-[(2S,4S,5S)-2-(4-methoxyphenyl)-5-methyl-1,3-dioxan-4-yl]-5,9-dimethyl-6-oxododeca-1,7-dien-1-yl}-N-methylformamide] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pereira, J.H, Petchprayoon, C, Moriarty, N.W, Fink, S.J, Cecere, G, Paterson, I, Adams, P.D, Marriott, G. | | Deposit date: | 2013-04-11 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical studies of actin in complex with synthetic macrolide tail analogues.
Chemmedchem, 9, 2014
|
|
3KFK
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (6.003 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3KFE
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3KFB
 
 | | Crystal structure of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3MMW
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
3MMU
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase, NICKEL (II) ION | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
6VAP
 
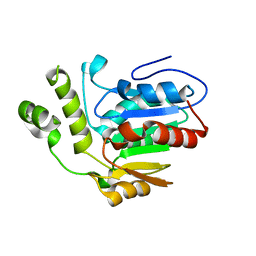 | | Structure of the type II thioesterase BorB from the borrelidin biosynthetic cluster | | Descriptor: | Thioesterase | | Authors: | Pereira, J.H, Curran, S.C, Baluyot, M.-J, Lake, J, Putz, H, Rosenburg, D, Keasling, J, Adams, P.D. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and Function of BorB, the Type II Thioesterase from the Borrelidin Biosynthetic Gene Cluster.
Biochemistry, 59, 2020
|
|
6URA
 
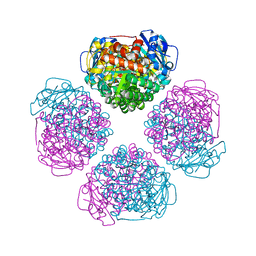 | | Crystal structure of RUBISCO from Promineofilum breve | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain | | Authors: | Pereira, J.H, Banda, D.M, Liu, A.K, Shih, P.M, Adams, P.D. | | Deposit date: | 2019-10-23 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel bacterial clade reveals origin of form I Rubisco.
Nat.Plants, 6, 2020
|
|
6W1H
 
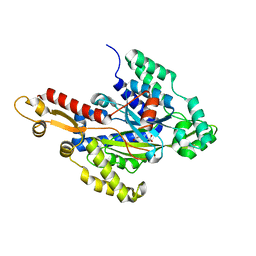 | | Crystal structure of the hydroxyglutarate synthase in complex with 2-oxoadipate from Pseudomonas putida | | Descriptor: | 2-OXOADIPIC ACID, Hydroxyglutarate synthase, NICKEL (II) ION | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
6W1G
 
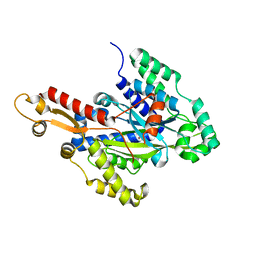 | | Crystal structure of the hydroxyglutarate synthase from Pseudomonas putida | | Descriptor: | Hydroxyglutarate synthase, NICKEL (II) ION | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
6W1K
 
 | | Crystal structure of the hydroxyglutarate synthase in complex with 2-oxoadipate from Oryza sativa | | Descriptor: | 2-OXOADIPIC ACID, Hydroxyglutarate synthase, NICKEL (II) ION, ... | | Authors: | Pereira, J.H, Thompson, M.G, Blake-Hedges, J.M, Keasling, J.D, Adams, P.D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An iron (II) dependent oxygenase performs the last missing step of plant lysine catabolism.
Nat Commun, 11, 2020
|
|
3D1N
 
 | | Structure of human Brn-5 transcription factor in complex with corticotrophin-releasing hormone gene promoter | | Descriptor: | 5'-D(*DAP*DGP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DA)-3', 5'-D(*DTP*DTP*DAP*DTP*DTP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DCP*DT)-3', POU domain, ... | | Authors: | Pereira, J.H, Ha, S.C, Kim, S.-H. | | Deposit date: | 2008-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human Brn-5 transcription factor in complex with CRH gene promoter.
J.Struct.Biol., 167, 2009
|
|
8D3Z
 
 | | Crystal structure of GalS1 in complex with Manganese from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactan synthase, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
