6RML
 
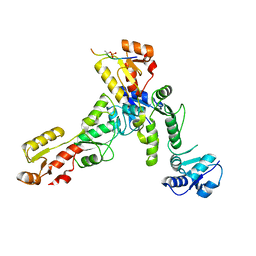 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | Descriptor: | 53BP1, DNA topoisomerase 2-binding protein 1 | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
6RMM
 
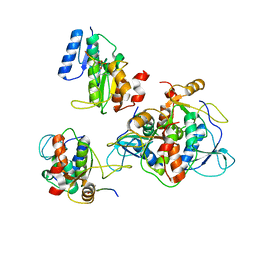 | |
6HM4
 
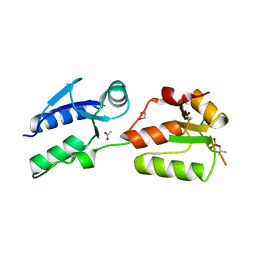 | | Crystal structure of Rad4 BRCT1,2 in complex with a Mdb1 phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA damage response protein Mdb1, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.770186 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
6HM3
 
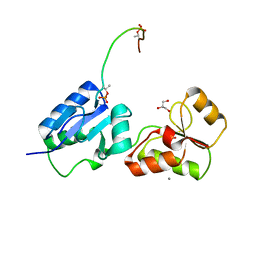 | | Crystal structure of Rad4 BRCT1,2 in complex with a Sld3 phosphopeptide | | Descriptor: | CALCIUM ION, DNA replication regulator sld3, GLYCEROL, ... | | Authors: | Day, M, Rappas, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2018-09-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77263618 Å) | | Cite: | BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.
Elife, 7, 2018
|
|
1AH8
 
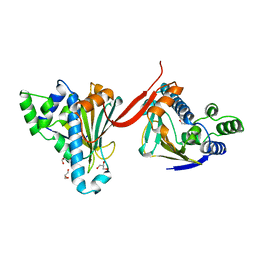 | |
1AH6
 
 | |
1HK7
 
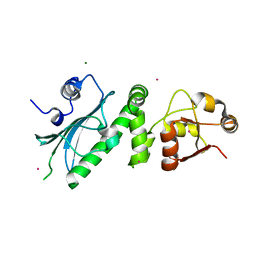 | | Middle Domain of HSP90 | | Descriptor: | CADMIUM ION, HEAT SHOCK PROTEIN HSP82, MAGNESIUM ION | | Authors: | Meyer, P, Prodromou, C, Roe, S.M, Pearl, L.H. | | Deposit date: | 2003-03-06 | | Release date: | 2004-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Analysis of the Middle Segment of Hsp90. Implications for ATP Hydrolysis and Client Protein and Cochaperone Interactions
Mol.Cell, 11, 2003
|
|
1KCF
 
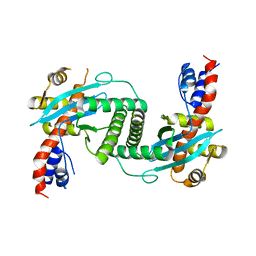 | | Crystal Structure of the Yeast Mitochondrial Holliday Junction Resolvase, Ydc2 | | Descriptor: | HYPOTHETICAL 30.2 KD PROTEIN C25G10.02 IN CHROMOSOME I, SULFATE ION | | Authors: | Ceschini, S, Keeley, A, McAlister, M.S.B, Oram, M, Phelan, J, Pearl, L.H, Tsaneva, I.R, Barrett, T.E. | | Deposit date: | 2001-11-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2.
EMBO J., 20, 2001
|
|
1KO9
 
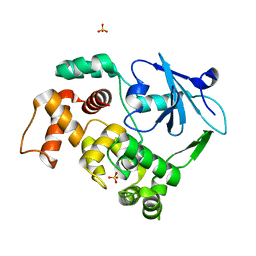 | | Native Structure of the Human 8-oxoguanine DNA Glycosylase hOGG1 | | Descriptor: | 8-oxoguanine DNA glycosylase, SULFATE ION | | Authors: | Bjoras, M, Seeberg, E, Luna, L, Pearl, L.H, Barrett, T.E. | | Deposit date: | 2001-12-20 | | Release date: | 2002-01-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase.
J.Mol.Biol., 317, 2002
|
|
1UV5
 
 | | GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH 6-BROMOINDIRUBIN-3'-OXIME | | Descriptor: | 6-BROMOINDIRUBIN-3'-OXIME, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Dajani, R, Pearl, L.H, Roe, S.M. | | Deposit date: | 2004-01-14 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gsk-3-Selective Inhibitors Derived from Tyrian Purple Indurubins
Chem.Biol., 10, 2003
|
|
2CE9
 
 | |
2IWS
 
 | | Radicicol analogues bound to the ATP site of HSP90 | | Descriptor: | (5Z)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
2IWX
 
 | | Analogues of radicicol bound to the ATP-binding site of Hsp90. | | Descriptor: | (5E)-14-CHLORO-15,17-DIHYDROXY-4,7,8,9,10,11-HEXAHYDRO-2-BENZOXACYCLOPENTADECINE-1,12(3H,13H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
2CE8
 
 | |
2IZO
 
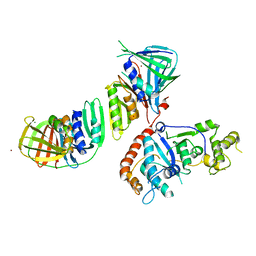 | | Structure of an Archaeal PCNA1-PCNA2-FEN1 Complex | | Descriptor: | DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C, FLAP STRUCTURE-SPECIFIC ENDONUCLEASE, ... | | Authors: | Dore, A.S, Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of an Archaeal PCNA1-PCNA2-Fen1 Complex: Elucidating PCNA Subunit and Client Enzyme Specificity.
Nucleic Acids Res., 34, 2006
|
|
2IWU
 
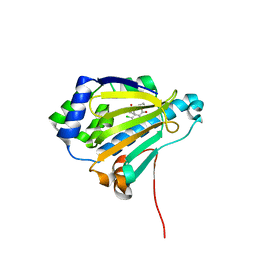 | | Analogues of radicicol bound to the ATP-binding site of Hsp90 | | Descriptor: | (5E)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
1USU
 
 | |
1US7
 
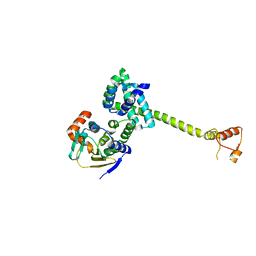 | | Complex of Hsp90 and P50 | | Descriptor: | HEAT SHOCK PROTEIN HSP82, HSP90 CO-CHAPERONE CDC37 | | Authors: | Roe, S.M, Ali, M.M.U, Pearl, L.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Hsp90 Regulation by the Protein Kinase-Specific Cochaperone p50(Cdc37)
Cell(Cambridge,Mass.), 116, 2004
|
|
1USV
 
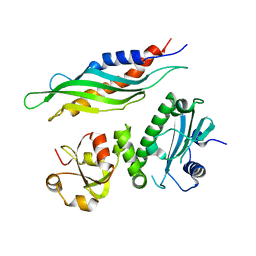 | |
1UNN
 
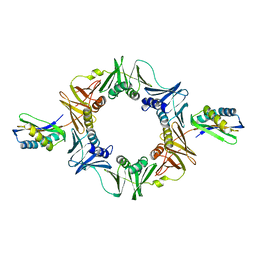 | |
2JKI
 
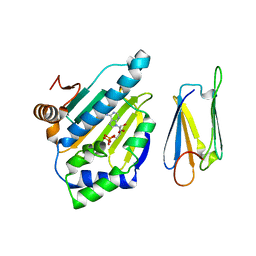 | | Complex of Hsp90 N-terminal and Sgt1 CS domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CYTOSOLIC HEAT SHOCK PROTEIN 90, SGT1-LIKE PROTEIN | | Authors: | Zhang, M, Pearl, L.H. | | Deposit date: | 2008-08-28 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Coupling of Hsp90- and Sgt1-Centred Multi-Protein Complexes.
Embo J., 27, 2008
|
|
1A4H
 
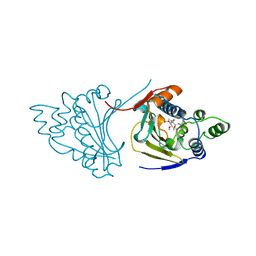 | |
1GEC
 
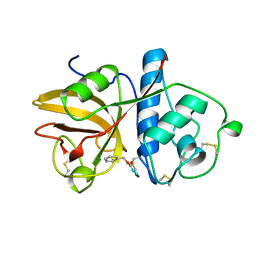 | | GLYCYL ENDOPEPTIDASE-COMPLEX WITH BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE COVALENTLY BOUND TO CYSTEINE 25 | | Descriptor: | BENZYLOXYCARBONYL-LEUCINE-VALINE-GLYCINE-METHYLENE INHIBITOR, GLYCYL ENDOPEPTIDASE | | Authors: | Ohara, B.P, Hemmings, A.M, Buttle, D.J, Pearl, L.H. | | Deposit date: | 1995-05-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
Biochemistry, 34, 1995
|
|
1GML
 
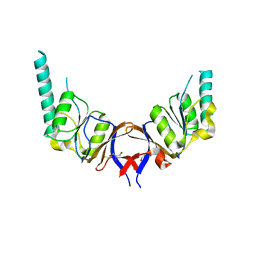 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | Descriptor: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-09-17 | | Release date: | 2002-06-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
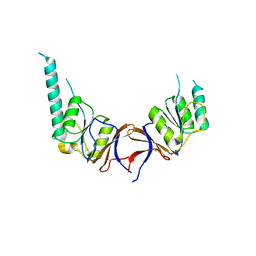 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | Descriptor: | CALCIUM ION, CCT-GAMMA | | Authors: | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | Deposit date: | 2001-10-01 | | Release date: | 2002-06-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
