1GTU
 
 | |
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
1YJ6
 
 | |
1YKC
 
 | |
1Y0G
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN, STRUCTURAL GENOMICS | | Descriptor: | 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, Protein yceI | | Authors: | Patskovsky, Y.V, Strokopytov, B, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PERIPLASMIC PROTEIN
To be Published
|
|
4GTU
 
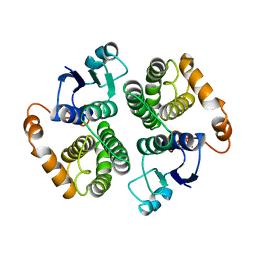 | |
3LOC
 
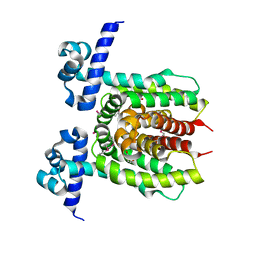 | | Crystal structure of putative transcriptional regulator ycdc | | Descriptor: | HTH-type transcriptional regulator rutR, URACIL | | Authors: | Patskovsky, Y.V, Knapik, A.A, Mennella, V, Burley, S.K, Minor, W, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Hypothetical Transcriptional Regulator Ycdc from Escherichia Coli
To be Published
|
|
3GTU
 
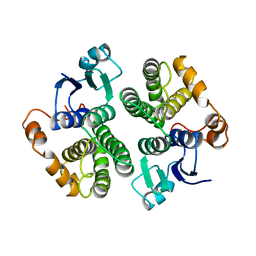 | |
1NEJ
 
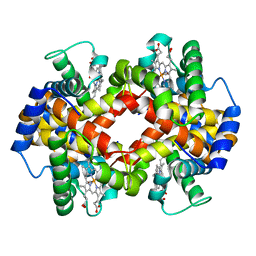 | | Crystalline Human Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Exhibits The R2 Quaternary State At Neutral pH In The Presence Of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-12-11 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1M9P
 
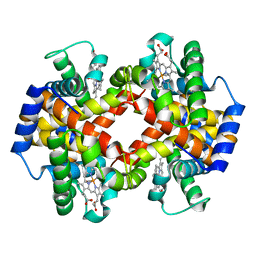 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XW5
 
 | |
2GTU
 
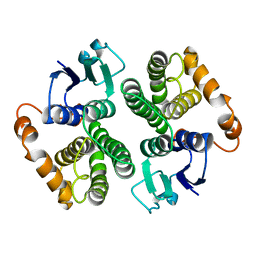 | | LIGAND-FREE HUMAN GLUTATHIONE S-TRANSFERASE M2-2 (E.C.2.5.1.18), MONOCLINIC CRYSTAL FORM | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Patskovska, L.N, Fedorov, A.A, Patskovsky, Y.V, Almo, S.C, Listowsky, I. | | Deposit date: | 1998-05-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The enhanced affinity for thiolate anion and activation of enzyme-bound glutathione is governed by an arginine residue of human Mu class glutathione S-transferases.
J.Biol.Chem., 275, 2000
|
|
2A1V
 
 | |
2GPY
 
 | | Crystal structure of putative O-methyltransferase from Bacillus halodurans | | Descriptor: | MAGNESIUM ION, O-methyltransferase, ZINC ION | | Authors: | Ramagopal, U.A, Patskovsky, Y.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-06 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative O-methyltransferase from Bacillus halodurans
To be Published
|
|
2HJ1
 
 | | Crystal structure of a 3D domain-swapped dimer of protein HI0395 from Haemophilus influenzae | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Ramagopal, U.A, Patskovsky, Y.V, Toro, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a 3D domain-swapped dimer of hypothetical protein from Haemophilus influenzae (CASP Target)
To be Published
|
|
