3TOU
 
 | | Crystal structure of GLUTATHIONE TRANSFERASE (TARGET EFI-501058) from Ralstonia solanacearum GMI1000 with GSH bound | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of GLUTATHIONE S-TRANSFERASE from Ralstonia solanacearum
To be Published
|
|
3IPL
 
 | | CRYSTAL STRUCTURE OF o-succinylbenzoic acid-CoA ligase FROM Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 2-succinylbenzoate--CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of o-succinylbenzoic acid-CoA ligase from Staphylococcus aureus
To be Published
|
|
3TOY
 
 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | Descriptor: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
3TWB
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and gluconic acid | | Descriptor: | CHLORIDE ION, D-gluconic acid, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3FHL
 
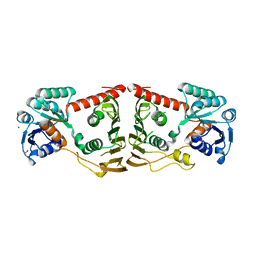 | | Crystal structure of a putative oxidoreductase from bacteroides fragilis nctc 9343 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative oxidoreductase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-09 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase from Bacteroides Fragilis Nctc 9343
To be Published
|
|
3IVR
 
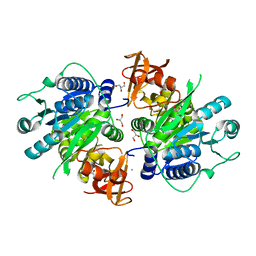 | | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA ligase FROM Rhodopseudomonas palustris CGA009 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative long-chain-fatty-acid CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA SYNTHASE FROM Rhodopseudomonas palustris CGA009
To be Published
|
|
3KEW
 
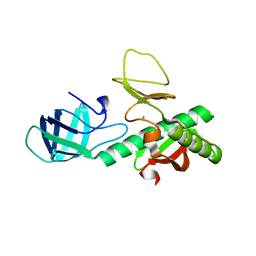 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
3KSU
 
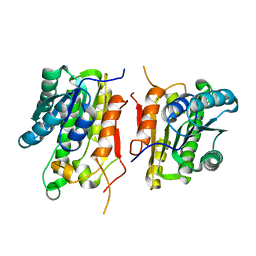 | | Crystal structure of short-chain dehydrogenase from oenococcus oeni psu-1 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase from Oenococcus Oeni Psu-1
To be Published
|
|
3L49
 
 | | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER SUBUNIT FROM Rhodobacter sphaeroides 2.4.1 | | Descriptor: | ABC sugar (Ribose) transporter, periplasmic substrate-binding subunit, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Do, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-05 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ABC SUGAR TRANSPORTER FROM Rhodobacter sphaeroides
To be Published
|
|
3LOM
 
 | | CRYSTAL STRUCTURE OF GERANYLTRANSFERASE FROM Legionella pneumophila | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LLW
 
 | | Crystal structure of geranyltransferase from helicobacter pylori 26695 | | Descriptor: | Geranyltranstransferase (IspA), SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Geranyltransferase from Helicobacter Pylori
To be Published
|
|
3LQ7
 
 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
3LMD
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum atcc 13032 | | Descriptor: | Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Ho, M, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Corynebacterium Glutamicum
To be Published
|
|
3LTE
 
 | | CRYSTAL STRUCTURE OF RESPONSE REGULATOR (SIGNAL RECEIVER DOMAIN) FROM Bermanella marisrubri | | Descriptor: | GLYCEROL, PHOSPHATE ION, Response regulator | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-15 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RESPONSE REGULATOR SIGNAL RECEIVER DOMAIN FROM Bermanella marisrubri RED65
To be Published
|
|
3LVS
 
 | | Crystal structure of farnesyl diphosphate synthase from rhodobacter capsulatus sb1003 | | Descriptor: | FARNESYL DIPHOSPHATE SYNTHASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3JU2
 
 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3K2W
 
 | | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica T6c | | Descriptor: | ACETATE ION, Betaine-aldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica
To be Published
|
|
3JVD
 
 | | Crystal structure of putative transcription regulation repressor (LACI family) FROM Corynebacterium glutamicum | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulators | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Foti, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-16 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LACI family protein from Corynebacterium glutamicum
To be Published
|
|
3K9D
 
 | | CRYSTAL STRUCTURE OF PROBABLE ALDEHYDE DEHYDROGENASE FROM Listeria monocytogenes EGD-e | | Descriptor: | ALDEHYDE DEHYDROGENASE, CHLORIDE ION, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase from Listeria Monocytogenes
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3MOG
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3KTS
 
 | | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365 | | Descriptor: | Glycerol uptake operon antiterminator regulatory protein, UNKNOWN LIGAND | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | CRYSTAL STRUCTURE OF GLYCEROL UPTAKE OPERON ANTITERMINATOR REGULATORY PROTEIN FROM LISTERIA MONOCYTOGENES STR. 4b F2365
To be Published
|
|
3MAE
 
 | | CRYSTAL STRUCTURE OF PROBABLE DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365 | | Descriptor: | 2-oxoisovalerate dehydrogenase E2 component, dihydrolipoamide acetyltransferase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A CATALYTIC DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM LISTERIA MONOCYTOGENES 4b F2365
To be Published
|
|
3MGK
 
 | | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum ATCC 824 | | Descriptor: | Intracellular protease/amidase related enzyme (ThiJ family) | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-06 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PROBABLE PROTEASE/AMIDASE FROM Clostridium acetobutylicum
To be Published
|
|
