3BE7
 
 | | Crystal structure of Zn-dependent arginine carboxypeptidase | | Descriptor: | ARGININE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Freeman, J, Iizuka, M, Bain, K, Rodgers, L, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-16 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional identification of incorrectly annotated prolidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
3BJS
 
 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
1XW6
 
 | | 1.9 angstrom resolution structure of human glutathione S-transferase M1A-1A complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-10-29 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1XWK
 
 | | 2.3 angstrom resolution crystal structure of human glutathione S-transferase M1A-1A complexed with glutathionyl-S-dinitrobenzene | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-11-01 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1YKC
 
 | |
1YJ6
 
 | |
2QUP
 
 | | Crystal structure of uncharacterized protein BH1478 from Bacillus halodurans | | Descriptor: | BH1478 protein, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Mckenzie, C, Bain, K.T, Smith, D, Ozyurt, S, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-06 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Bh1478 from Bacillus Halodurans.
To be Published
|
|
2QH5
 
 | | Crystal structure of mannose-6-phosphate isomerase from Helicobacter pylori | | Descriptor: | Mannose-6-phosphate isomerase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Iizuka, M, Maletic, M, Wasserman, S.R, Koss, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mannose-6-phosphate isomerase from Helicobacter pylori.
To be Published
|
|
2QSD
 
 | | Crystal structure of a protein Il1583 from Idiomarina loihiensis | | Descriptor: | GLYCEROL, Uncharacterized conserved protein | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Romero, R, Rutter, M, Koss, J, Mckenzie, C, Gheyi, T, Bain, K, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Protein Il1583 from Idiomarina loihiensis.
To be Published
|
|
2QV5
 
 | | Crystal structure of uncharacterized protein ATU2773 from Agrobacterium tumefaciens C58 | | Descriptor: | GLYCEROL, Uncharacterized protein Atu2773 | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Dickey, M, Bain, K.T, Iizuka, M, Smith, D, Rodgers, L, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-07 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Atu2773 from Agrobacterium Tumefaciens.
To be Published
|
|
1YIF
 
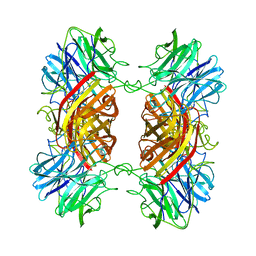 | | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | BETA-1,4-XYLOSIDASE | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS
To be Published
|
|
1YZY
 
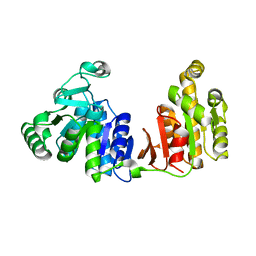 | |
1Y0G
 
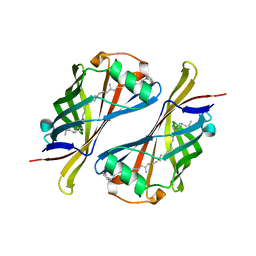 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN, STRUCTURAL GENOMICS | | Descriptor: | 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, Protein yceI | | Authors: | Patskovsky, Y.V, Strokopytov, B, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PERIPLASMIC PROTEIN
To be Published
|
|
1Z2I
 
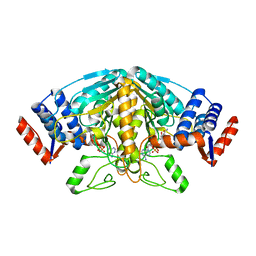 | |
1Y1L
 
 | |
1YGA
 
 | |
2R1F
 
 | | Crystal structure of predicted aminodeoxychorismate lyase from Escherichia coli | | Descriptor: | CADMIUM ION, GLYCEROL, Predicted aminodeoxychorismate lyase, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Rutter, M, Lau, C, Maletic, M, Smith, D, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Predicted Aminodeoxychorismate Lyase from Escherichia coli.
To be Published
|
|
2RDM
 
 | | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Yan, Q, Zhan, C, Toro, R, Meyer, A.J, Gilmore, M, Hu, S, Groshong, C, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of response regulator receiver protein from Sinorhizobium medicae WSM419.
To be Published
|
|
2F3M
 
 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
2RDX
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Patskovsky, Y, Bonanno, J, Sauder, J.M, Ozyurt, S, Gilmore, M, Lau, C, Maletic, M, Gheyi, T, Wasserman, S.R, Koss, J, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM.
To be Published
|
|
4NHW
 
 | | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275, with one glutathione bound per one protein subunit | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275
To be Published
|
|
4Q60
 
 | | Crystal structure of a 4-hydroxyproline epimerase from Burkholderia Multivorans atcc 17616, target EFI-506586, open form, with bound pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PROLINE RACEMASE, PYRROLE-2-CARBOXYLATE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF PROLINE RACEMASE Bmul_4447 FROM Burkholderia multivorans, TARGET EFI-506586
To be Published
|
|
4MPG
 
 | | Crystal structure of human glutathione transferase theta-2, complex with glutathione and unknown ligand, target EFI-507257 | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase theta-2, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target EFI-507257)
To be Published
|
|
4PX1
 
 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
