6UFI
 
 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
6TZP
 
 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
2KTM
 
 | | Solution NMR structure of H2H3 domain of ovine prion protein (residues 167-234) | | Descriptor: | Major prion protein | | Authors: | Pastore, A, Adrover, M, Pauwels, K, de Chiara, C, Prigent, S, Rezeai, H. | | Deposit date: | 2010-02-04 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Prion fibrillization is mediated by a native structural element that comprises helices H2 and H3.
J.Biol.Chem., 285, 2010
|
|
7QAB
 
 | |
2LFU
 
 | | The structure of a N. meningitides protein targeted for vaccine development | | Descriptor: | Gna2132 | | Authors: | Esposito, V, Musi, V, De Chiara, C, Kelly, G, Veggi, D, Pizza, M, Pastore, A. | | Deposit date: | 2011-07-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal Domain of Neisseria Heparin Binding Antigen (NHBA), One of the Main Antigens of a Novel Vaccine against Neisseria meningitidis.
J.Biol.Chem., 286, 2011
|
|
1VIH
 
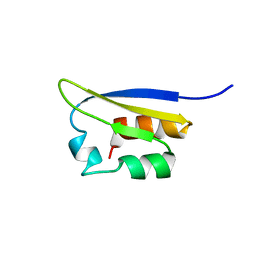 | | NMR STUDY OF VIGILIN, REPEAT 6, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
1VIG
 
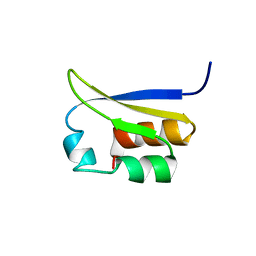 | | NMR STUDY OF VIGILIN, REPEAT 6, 40 STRUCTURES | | Descriptor: | VIGILIN | | Authors: | Musco, G, Stier, G, Joseph, C, Morelli, M.A.C, Nilges, M, Gibson, T.J, Pastore, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome.
Cell(Cambridge,Mass.), 85, 1996
|
|
2BZT
 
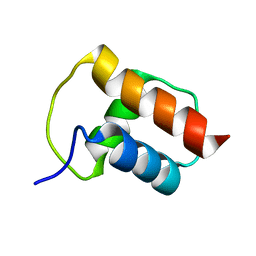 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
1H8B
 
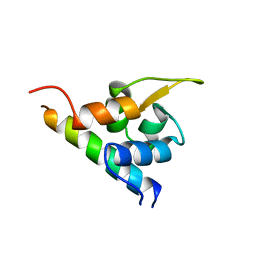 | | EF-hands 3,4 from alpha-actinin / Z-repeat 7 from titin | | Descriptor: | ALPHA-ACTININ 2, SKELETAL MUSCLE ISOFORM, TITIN | | Authors: | Atkinson, R.A, Joseph, C, Kelly, G, Muskett, F.W, Frenkiel, T.A, Nietlispach, D, Pastore, A. | | Deposit date: | 2001-02-01 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ca2+-Independent Binding of an EF-Hand Domain to a Novel Motif in the Alpha-Actinin-Titin Complex
Nat.Struct.Biol., 8, 2001
|
|
8Q5Q
 
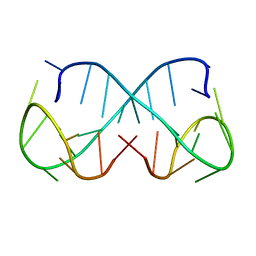 | | d(ATTTC)3 dimeric structure | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*(DNR)P*AP*TP*TP*TP*CP*AP*TP*TP*TP*C)-3') | | Authors: | Trajkovski, M, Pastore, A, Plavec, J. | | Deposit date: | 2023-08-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Dimeric structures of DNA ATTTC repeats promoted by divalent cations.
Nucleic Acids Res., 52, 2024
|
|
1LY7
 
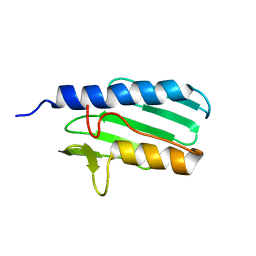 | | The solution structure of the the c-terminal domain of frataxin, the protein responsible for friedreich ataxia | | Descriptor: | frataxin | | Authors: | Musco, G, Stier, G, Kolmerer, B, Adinolfi, S, Martin, S, Frenkiel, T, Gibson, T, Pastore, A. | | Deposit date: | 2002-06-07 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Towards a structural understanding of Friedreich's
ataxia: the solution structure of frataxin
Structure Fold.Des., 8, 2000
|
|
1SOY
 
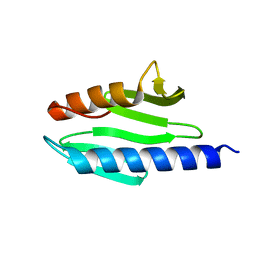 | | Solution structure of the bacterial frataxin orthologue, CyaY | | Descriptor: | CyaY protein | | Authors: | Nair, M, Adinolfi, S, Pastore, C, Kelly, G, Temussi, P, Pastore, A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Frataxin Ortholog, CyaY; Mapping the Iron Binding Sites
Structure, 12, 2004
|
|
4AQP
 
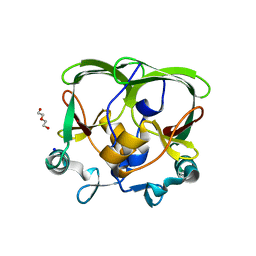 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
4APT
 
 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
1YZB
 
 | | Solution structure of the Josephin domain of Ataxin-3 | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Nicastro, G, Masino, L, Menon, R.P, Knowles, P.P, McDonald, N.Q, Pastore, A. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Josephin domain of ataxin-3: Structural determinants for molecular recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5LSD
 
 | | recombinant mouse Nerve Growth Factor | | Descriptor: | Beta-nerve growth factor | | Authors: | Paoletti, F, de Chiara, C, Kelly, G, Lamba, D, Cattaneo, A, Pastore, A. | | Deposit date: | 2016-08-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conformational Rigidity within Plasticity Promotes Differential Target Recognition of Nerve Growth Factor.
Front Mol Biosci, 3, 2016
|
|
1NCT
 
 | | TITIN MODULE M5, N-TERMINALLY EXTENDED, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1NCU
 
 | | Titin Module M5, N-terminally Extended, NMR | | Descriptor: | TITIN | | Authors: | Pfuhl, M, Pastore, A. | | Deposit date: | 1996-08-13 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | When a module is also a domain: the role of the N terminus in the stability and the dynamics of immunoglobulin domains from titin.
J.Mol.Biol., 265, 1997
|
|
1ZTA
 
 | | THE SOLUTION STRUCTURE OF A LEUCINE-ZIPPER MOTIF PEPTIDE | | Descriptor: | LEUCINE ZIPPER MONOMER | | Authors: | Saudek, V, Pastore, A, Castiglione Morelli, M.A, Frank, R, Gausepohl, H, Gibson, T. | | Deposit date: | 1990-10-11 | | Release date: | 1993-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a leucine-zipper motif peptide.
Protein Eng., 4, 1991
|
|
6HRJ
 
 | | Apo - YndL | | Descriptor: | CITRATE ANION, SULFATE ION, YndL, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
6HRI
 
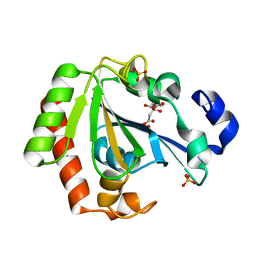 | | Native YndL | | Descriptor: | CITRATE ANION, IMIDAZOLE, SULFATE ION, ... | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
FEBS J., 285, 2018
|
|
5ONJ
 
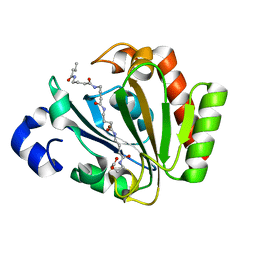 | | YnDL in Complex with 5 amino acid (PGA) complex | | Descriptor: | (2R,3S,4R,5R,6R)-6-((1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYLOXY)-5-AMINO-2-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, YndL, ZINC ION | | Authors: | Ramaswamy, S, Rasheed, M, Morelli, C, Calvio, C, Sutton, B, Pastore, A. | | Deposit date: | 2017-08-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of PghL hydrolase bound to its substrate poly-gamma-glutamate.
Febs J., 285, 2018
|
|
2M41
 
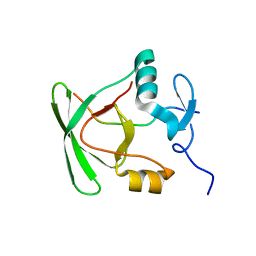 | |
6FCO
 
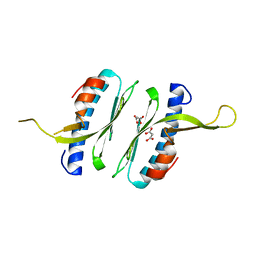 | | Structural and functional characterisation of Frataxin (FXN) like protein from Chaetomium thermophilum | | Descriptor: | MALONIC ACID, Mitochondrial frataxin-like protein | | Authors: | Jamshidiha, M, Rasheed, M, Pastore, A, Cota, E. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and functional characterization of a frataxin from a thermophilic organism.
FEBS J., 286, 2019
|
|
2BTT
 
 | |
