6PDC
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 42 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-[(prop-2-en-1-yl)oxy]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDB
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 80 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-(pyrimidin-2-yl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDD
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 41 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-[(propan-2-yl)oxy]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6Q4Y
 
 | | Structure of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana, in complex with mannose | | Descriptor: | LmxM MPT-2, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q4X
 
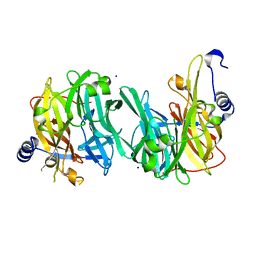 | | Structure of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q4W
 
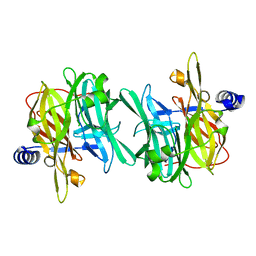 | | Structure of MPT-1, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana | | Descriptor: | LmxM MPT-1 | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q4Z
 
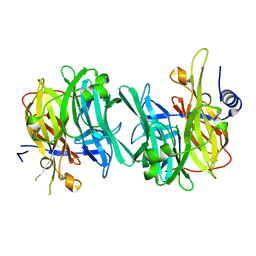 | | Structure of an inactive variant (D94N) of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana, in complex with beta-1,2-mannobiose | | Descriptor: | LmxM MPT-2 D94N, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Kloehn, J, Viera-Lara, M, Stanton, L, Cobbold, S, Pires, D.E, Hanssen, E, Parker, M.W, Ascher, D.B, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6OIP
 
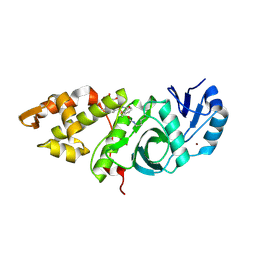 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 34 | | Descriptor: | 2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6Q50
 
 | | Structure of MPT-4, a mannose phosphorylase from Leishmania mexicana, in complex with phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, MPT-4, PHOSPHATE ION, ... | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6OIQ
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 63 | | Descriptor: | 2-fluoro-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6OIO
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 60 | | Descriptor: | Histone acetyltransferase KAT8, N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, ZINC ION | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6OWH
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 92 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
1OWT
 
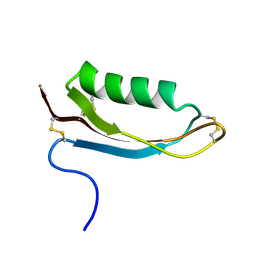 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1PFO
 
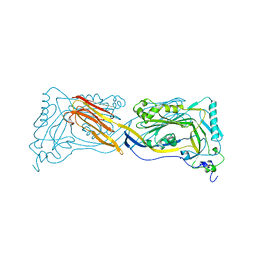 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
1PX6
 
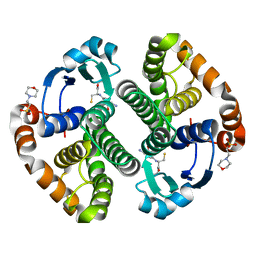 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1MD3
 
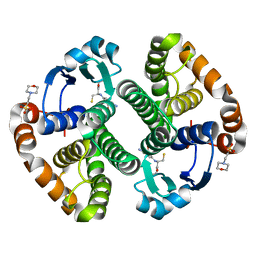 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to alanine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
1PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-03-23 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1PX7
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to glutamate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1MWP
 
 | | N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN | | Descriptor: | AMYLOID A4 PROTEIN | | Authors: | Rossjohn, J, Cappai, R, Feil, S.C, Henry, A, McKinstry, W.J, Galatis, D, Hesse, L, Multhaup, G, Beyreuther, K, Masters, C.L, Parker, M.W. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein.
Nat.Struct.Biol., 6, 1999
|
|
3VQ4
 
 | | Fragments bound to HIV-1 integrase | | Descriptor: | (5-phenyl-1,2-oxazol-3-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQ6
 
 | | HIV-1 IN core domain in complex with (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol | | Descriptor: | (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
6VKA
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | 2,2'-{ethane-1,2-diylbis[oxyethane-2,1-diylcarbamoyl-4,1-phenyleneethyne-2,1-diyl(5-methyl-1-benzofuran-2,3-diyl)]}diacetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | HIV Integrase core domain (IN) in complex with dimeric spanning inhibitor
To Be Published
|
|
6VLH
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-01-24 | | Release date: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | HIV Integrase core domain (IN) in complex with dimeric spanning inhibitor
To Be Published
|
|
3VQC
 
 | | HIV-1 IN core domain in complex with (5-METHYL-3-PHENYL-1,2-OXAZOL-4-YL)METHANOL | | Descriptor: | (5-methyl-3-phenyl-1,2-oxazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQE
 
 | | HIV-1 IN core domain in complex with [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
