2HP7
 
 | | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor | | Descriptor: | Flagellar motor switch protein FliM | | Authors: | Crane, B.R, Park, S, Lowder, B, Bilwes, A.M, Blair, D.F. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4JOU
 
 | |
4JNJ
 
 | | Structure based engineering of streptavidin monomer with a reduced biotin dissociation rate | | Descriptor: | BIOTIN, Streptavidin/Rhizavidin Hybrid, ZINC ION | | Authors: | DeMonte, D, Drake, E.J, Hong Lim, K, Gulick, A.M, Park, S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure-based engineering of streptavidin monomer with a reduced biotin dissociation rate.
Proteins, 81, 2013
|
|
6II6
 
 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O in complex with a human ADP-ribosylation factor 3 (ARF3) | | Descriptor: | ADP-ribosylation factor 3, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6II2
 
 | | Crystal structure of alpha-beta hydrolase (ABH) and Makes Caterpillars Floppy (MCF)-Like effectors of Vibrio vulnificus MO6-24/O | | Descriptor: | Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyung, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6II0
 
 | | Crystal structure of the Makes Caterpillars Floppy (MCF)-Like effector of Vibrio vulnificus MO6-24/O | | Descriptor: | GLYCEROL, Putative RTX-toxin | | Authors: | Lee, Y, Kim, B.S, Choi, S, Lee, E.Y, Park, S, Hwang, J, Kwon, Y, Hyun, J, Lee, C, Eom, S.H, Kim, M.H. | | Deposit date: | 2018-10-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Makes caterpillars floppy-like effector-containing MARTX toxins require host ADP-ribosylation factor (ARF) proteins for systemic pathogenicity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2NC4
 
 | | Solution Structure of N-Galactosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-galactopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-L-idopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
5YX4
 
 | | Isoliquiritigenin-complexed Chalcone isomerase (S189A) from the Antarctic vascular plant Deschampsia Antarctica (DaCHI1) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Chalcone-flavonone isomerase family protein | | Authors: | Lee, C.W, Park, S, Lee, J.H. | | Deposit date: | 2017-12-01 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and enzymatic properties of chalcone isomerase from the Antarctic vascular plant Deschampsia antarctica Desv.
PLoS ONE, 13, 2018
|
|
5YX3
 
 | |
6A4E
 
 | |
6A4D
 
 | |
6A4A
 
 | |
6A4F
 
 | |
2NC3
 
 | | Solution Structure of N-Allosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-allopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC5
 
 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | Descriptor: | Pin1 WW Domain, beta-D-xylopyranose | | Authors: | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | Deposit date: | 2016-03-20 | | Release date: | 2016-06-08 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
5U9B
 
 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 in complex with human cardiac troponin T pre-mRNA | | Descriptor: | Muscleblind-like protein 1, RNA (5'-R(P*GP*UP*CP*UP*CP*GP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
5U6L
 
 | | Solution structure of the zinc fingers 3 and 4 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
5U6H
 
 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
2AIV
 
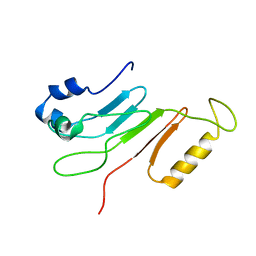 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
1J7N
 
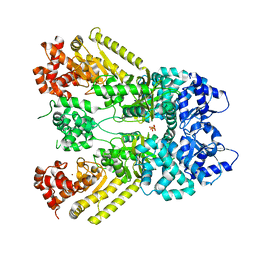 | | Anthrax Toxin Lethal factor | | Descriptor: | Lethal Factor precursor, SULFATE ION, ZINC ION | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-05-17 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
2JXB
 
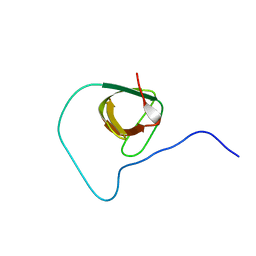 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
1L2Z
 
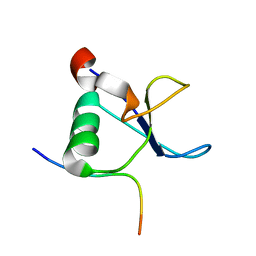 | | CD2BP2-GYF domain in complex with proline-rich CD2 tail segment peptide | | Descriptor: | CD2 ANTIGEN (CYTOPLASMIC TAIL)-BINDING PROTEIN 2, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Freund, C, Kuhne, R, Yang, H, Park, S, Reinherz, E.L, Wagner, G. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules
Embo J., 21, 2002
|
|
2KI2
 
 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
1JKY
 
 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
