7XZ3
 
 | | Crystal structure of the Type I-B CRISPR-associated protein, Csh2 from Thermobaculum terrenum | | Descriptor: | CRISPR-associated protein, Csh2 family | | Authors: | Seo, P.W, Gu, D.H, Park, S.Y, Kim, J.S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural characterization of the type I-B CRISPR Cas7 from Thermobaculum terrenum.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
1GEM
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450CAM, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
1GEK
 
 | | STRUCTURAL CHARACTERIZATION OF N-BUTYL-ISOCYANIDE COMPLEXES OF CYTOCHROMES P450NOR AND P450CAM | | Descriptor: | CYTOCHROME P450CAM, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, D.-S, Park, S.-Y, Yamane, K, Shiro, Y. | | Deposit date: | 2000-11-13 | | Release date: | 2000-12-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam.
Biochemistry, 40, 2001
|
|
5Z2E
 
 | |
5Z2F
 
 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
5Z2D
 
 | |
1GDV
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3QJN
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
6UUP
 
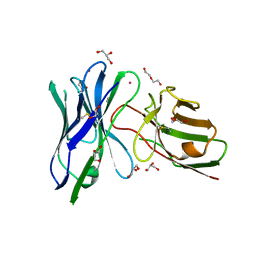 | | Structure of anti-hCD33 conditional scFv | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-CD33 conditional scFv, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kimberlin, C.R, Park, S. | | Deposit date: | 2019-10-31 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.20001864 Å) | | Cite: | Direct control of CAR T cells through small molecule-regulated antibodies.
Nat Commun, 12, 2021
|
|
1EHG
 
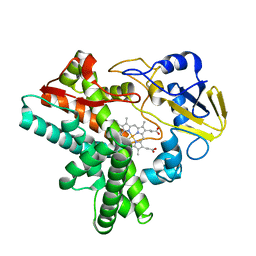 | |
1EHF
 
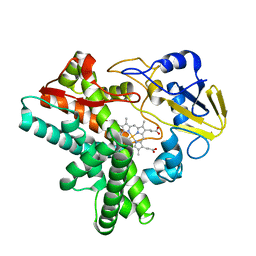 | |
1EHE
 
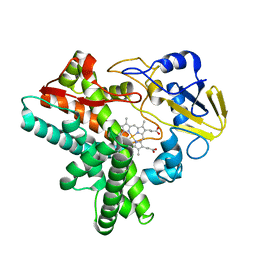 | |
7DP1
 
 | |
7DP0
 
 | |
7DP2
 
 | |
3QJM
 
 | | Structural flexibility of Shank PDZ domain is important for its binding to different ligands | | Descriptor: | Beta-PIX, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Lee, J.H, Park, H, Park, S.J, Kim, H.J, Eom, S.H. | | Deposit date: | 2011-01-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The structural flexibility of the shank1 PDZ domain is important for its binding to different ligands
Biochem.Biophys.Res.Commun., 407, 2011
|
|
6UY3
 
 | |
2B9L
 
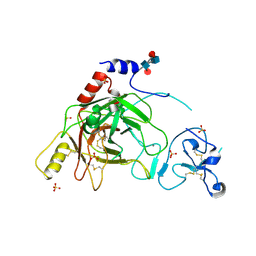 | | Crystal structure of prophenoloxidase activating factor-II from the beetle Holotrichia diomphalia | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piao, S, Song, Y.-L, Park, S.Y, Lee, B.L, Oh, B.-H, Ha, N.-C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a clip-domain serine protease and functional roles of the clip domains.
Embo J., 24, 2005
|
|
7XCB
 
 | |
4B04
 
 | | Crystal structure of the Catalytic Domain of Human DUSP26 (C152S) | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 26 | | Authors: | Won, E.-Y, Lee, D.Y, Park, S.G, Yokoyama, S, Kim, S.J, Chi, S.-W. | | Deposit date: | 2012-06-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | High-Resolution Crystal Structure of the Catalytic Domain of Human Dual-Specificity Phosphatase 26
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4O6W
 
 | | Peptide-Based Inhibitors of Plk1 Polo-box Domain | | Descriptor: | Peptide-Based inhibitor, Serine/threonine-protein kinase PLK1 | | Authors: | Qian, W.-J, Park, J.-E, Lim, D.C, Park, S.-Y, Lee, K.-W, Yaffe, M.B, Lee, K.S, Burke, T.R. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Mono-anionic phosphopeptides produced by unexpected histidine alkylation exhibit high plk1 polo-box domain-binding affinities and enhanced antiproliferative effects in hela cells.
Biopolymers, 102, 2014
|
|
1EW0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE SENSOR DOMAIN OF RMFIXL(FERROUS FORM) | | Descriptor: | FIXL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1QSI
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | CARBON MONOXIDE, PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
