3QYB
 
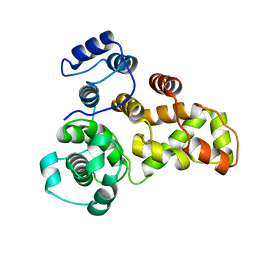 | |
3QYE
 
 | | Crystal Structure of Human TBC1D1 RabGAP domain | | Descriptor: | TBC1 domain family member 1 | | Authors: | Park, S.Y, Shoelson, S.E. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human TBC1D1 and TBC1D4 (AS160) RabGTPase-activating protein (RabGAP) domains reveal critical elements for GLUT4 translocation.
J.Biol.Chem., 286, 2011
|
|
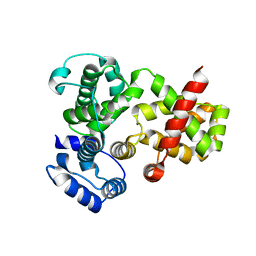
1UIW
 
 | | Crystal Structures of Unliganded and Half-Liganded Human Hemoglobin Derivatives Cross-Linked between Lys 82beta1 and Lys 82beta2 | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Park, S.-Y, Shibayama, N, Tame, J.R.H. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of unliganded and half-liganded human hemoglobin derivatives cross-linked between Lys 82beta1 and Lys 82beta2
Biochemistry, 43, 2004
|
|
2E47
 
 | |
5X12
 
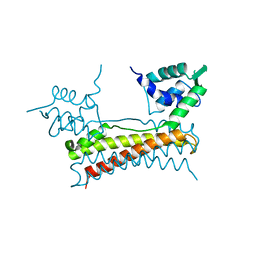 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
2KNH
 
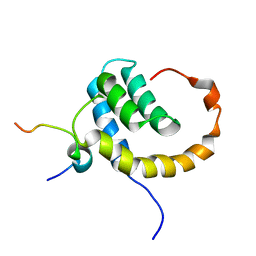 | | The Solution structure of the eTAFH domain of AML1-ETO complexed with HEB peptide | | Descriptor: | Protein CBFA2T1, Transcription factor 12 | | Authors: | Park, S, Cierpicki, T, Tonelli, M, Bushweller, J.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity.
Blood, 113, 2009
|
|
5X14
 
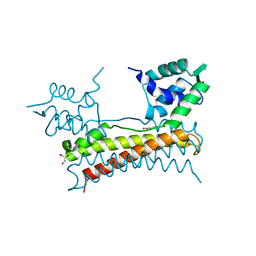 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
2KYU
 
 | | The solution structure of the PHD3 finger of MLL | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression.
Biochemistry, 49, 2010
|
|
7CBV
 
 | |
4OHT
 
 | | Crystal structure of succinic semialdehyde dehydrogenase from Streptococcus pyogenes in complex with NADP+ as the cofactor | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | Authors: | Park, S.A, Jang, E.H, Chi, Y.M, Lee, K.S. | | Deposit date: | 2014-01-18 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural Characterization for Cofactor Preference of Succinic Semialdehyde Dehydrogenase from Streptococcus pyogenes.
Mol.Cells, 37, 2014
|
|
1IRD
 
 | |
4QQA
 
 | |
4QQQ
 
 | |
1L5Z
 
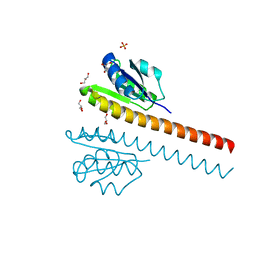 | | CRYSTAL STRUCTURE OF THE E121K SUBSTITUTION OF THE RECEIVER DOMAIN OF SINORHIZOBIUM MELILOTI DCTD | | Descriptor: | C4-DICARBOXYLATE TRANSPORT TRANSCRIPTIONAL REGULATORY PROTEIN DCTD, GLYCEROL, SULFATE ION | | Authors: | Park, S, Meyer, M, Jones, A.D, Yennawar, H.P, Yennawar, N.H, Nixon, B.T. | | Deposit date: | 2002-03-08 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain
FASEB J., 16, 2002
|
|
6KSY
 
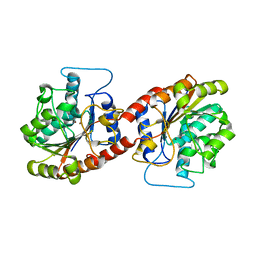 | |
6IY6
 
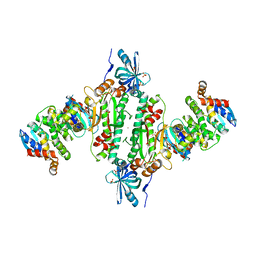 | | Crystal structure of human cytosolic aspartyl-tRNA synthetase (DRS) in complex with glutathion-S transferase (GST) domains from Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2) and glutamyl-prolyl-tRNA synthetase (EPRS) | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, Aspartate--tRNA ligase, cytoplasmic, ... | | Authors: | Park, S.H, Hahn, H, Han, B.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The DRS-AIMP2-EPRS subcomplex acts as a pivot in the multi-tRNA synthetase complex.
Iucrj, 6, 2019
|
|
5U7G
 
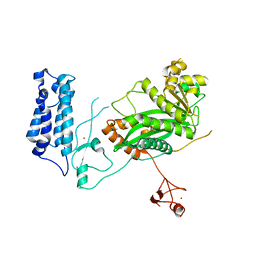 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3SFT
 
 | |
2KYX
 
 | | Solution structure of the RRM domain of CYP33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Park, S, Bushweller, J.H. | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression .
Biochemistry, 49, 2010
|
|
2CH4
 
 | |
2CH7
 
 | |
2B86
 
 | |
3T8Y
 
 | |
1U0S
 
 | | Chemotaxis kinase CheA P2 domain in complex with response regulator CheY from the thermophile thermotoga maritima | | Descriptor: | Chemotaxis protein cheA, Chemotaxis protein cheY | | Authors: | Park, S.Y, Beel, B.D, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-07-14 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1F26
 
 | |
