3BD4
 
 | |
3BD5
 
 | |
3BD3
 
 | |
4FVC
 
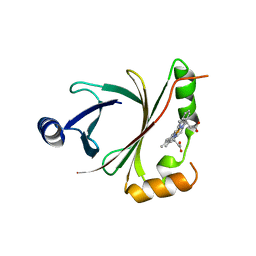 | | HmoB structure with heme | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein yhgC | | Authors: | Park, S. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of HmoB with Heme
To be Published
|
|
3H1T
 
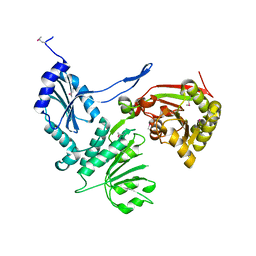 | |
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
2JPX
 
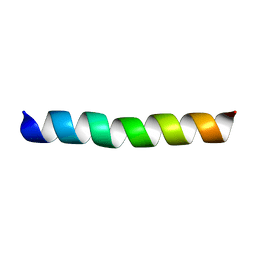 | | A18H Vpu TM structure in lipid bilayers | | Descriptor: | Vpu protein | | Authors: | Park, S, Opella, S.J. | | Deposit date: | 2007-05-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Conformational changes induced by a single amino acid substitution in the trans-membrane domain of Vpu: implications for HIV-1 susceptibility to channel blocking drugs
Protein Sci., 16, 2007
|
|
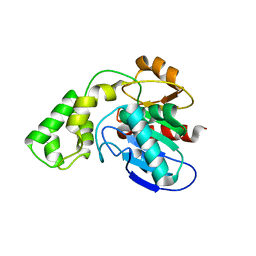
3BF7
 
 | |
3BF8
 
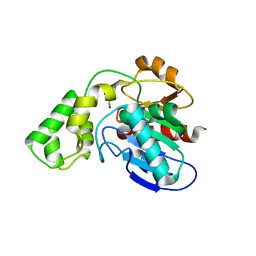 | |
2KSJ
 
 | | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | | Descriptor: | Capsid protein G8P | | Authors: | Park, S, Marassi, F, Black, D, Opella, S.J. | | Deposit date: | 2010-01-05 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR, SOLUTION NMR | | Cite: | Structure and dynamics of the membrane-bound form of Pf1 coat protein: implications of structural rearrangement for virus assembly.
Biophys.J., 99, 2010
|
|
1IO8
 
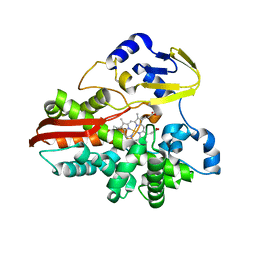 | | Thermophilic cytochrome P450 (CYP119) from sulfolobus solfataricus: High resolution structural origin of its thermostability and functional properties | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties
J.Inorg.Biochem., 91, 2002
|
|
1IO9
 
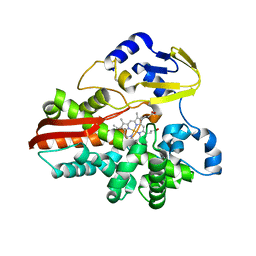 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
7MBM
 
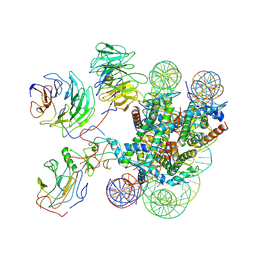 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode01 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
7MBN
 
 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode02 | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
1IO7
 
 | | THERMOPHILIC CYTOCHROME P450 (CYP119) FROM SULFOLOBUS SOLFATARICUS: HIGH RESOLUTION STRUCTURAL ORIGIN OF ITS THERMOSTABILITY AND FUNCTIONAL PROPERTIES | | Descriptor: | CYTOCHROME P450 CYP119, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yamane, K, Adachi, S, Shiro, Y, Sligar, S.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-02-08 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties.
J.Inorg.Biochem., 91, 2002
|
|
3WX7
 
 | | Crystal structure of COD | | Descriptor: | ACETATE ION, CALCIUM ION, Chitin oligosaccharide deacetylase, ... | | Authors: | Park, S.-Y, Sugiyama, K, Hirano, T, Nishio, T. | | Deposit date: | 2014-07-18 | | Release date: | 2014-12-03 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus.
FEBS Lett., 589, 2015
|
|
5J2O
 
 | |
3OZQ
 
 | |
6JL9
 
 | | Crystal structure of a frog ependymin related protein | | Descriptor: | CALCIUM ION, Ependymin-related 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JLD
 
 | | Crystal structure of a human ependymin related protein | | Descriptor: | Mammalian ependymin-related protein 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
7E8N
 
 | | Crystal structure of Type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554 | | Descriptor: | CITRIC ACID, Citrate synthase | | Authors: | Park, S.-H, Lee, C.W, Bae, D.-W, Lee, J.H. | | Deposit date: | 2021-03-02 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the cooperative activation of type II citrate synthase (HyCS) from Hymenobacter sp. PAMC 26554.
Int.J.Biol.Macromol., 183, 2021
|
|
1XKR
 
 | | X-ray Structure of Thermotoga maritima CheC | | Descriptor: | chemotaxis protein CheC | | Authors: | Park, S.Y, Chao, X, Gonzalez-Bonet, G, Beel, B.D, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-09-29 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Function of an Unusual Family of Protein Phosphatases; The Bacterial Chemotaxis Proteins CheC and CheX
Mol.Cell, 16, 2004
|
|
2LNL
 
 | | Structure of human CXCR1 in phospholipid bilayers | | Descriptor: | C-X-C chemokine receptor type 1 | | Authors: | Park, S, Das, B.B, Casagrande, F, Nothnagel, H, Chu, M, Kiefer, H, Maier, K, De Angelis, A, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-12-31 | | Release date: | 2012-10-17 | | Last modified: | 2016-04-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the chemokine receptor CXCR1 in phospholipid bilayers.
Nature, 491, 2012
|
|
1PI7
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
1PI8
 
 | | Structure of the channel-forming trans-membrane domain of Virus protein "u" (Vpu) from HIV-1 | | Descriptor: | VPU protein | | Authors: | Park, S.H, Mrse, A.A, Nevzorov, A.A, Mesleh, M.F, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Three-dimensional structure of the channel-forming trans-membrane domain of virus protein "u" (Vpu) from HIV-1
J.Mol.Biol., 333, 2003
|
|
