6LTZ
 
 | |
5JEB
 
 | |
5JV0
 
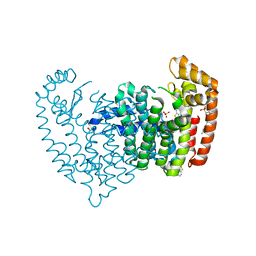 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-038 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-2-(3-fluorophenyl)-1-{[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
5JV1
 
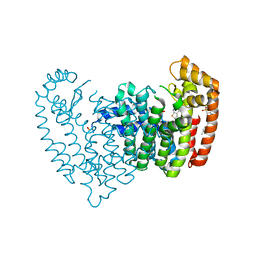 | | Crystal structure of human FPPS in complex with an allosteric inhibitor CL-08-066 | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION, [(1R)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-(3-fluorophenyl)ethyl]phosphonic acid | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-05-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacophore Mapping of Thienopyrimidine-Based Monophosphonate (ThP-MP) Inhibitors of the Human Farnesyl Pyrophosphate Synthase.
J. Med. Chem., 60, 2017
|
|
6OAH
 
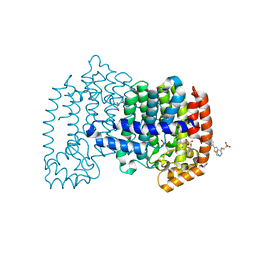 | |
6OAG
 
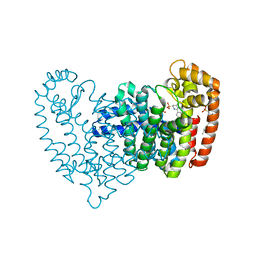 | | Crystal structure of human FPPS in complex with an allosteric inhibitor YF-02-82 | | Descriptor: | Farnesyl pyrophosphate synthase, PHOSPHATE ION, [(1S)-1-{[6-(3-chloro-4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Park, J, Berghuis, A.M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chirality-Driven Mode of Binding of alpha-Aminophosphonic Acid-Based Allosteric Inhibitors of the Human Farnesyl Pyrophosphate Synthase (hFPPS).
J.Med.Chem., 62, 2019
|
|
4DEM
 
 | | Crystal structure of human FPPS in complex with YS_04_70 | | Descriptor: | Farnesyl pyrophosphate synthase, MAGNESIUM ION, [({4-[4-(propan-2-yloxy)phenyl]pyridin-2-yl}amino)methanediyl]bis(phosphonic acid) | | Authors: | Park, J, Lin, Y.-S, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2012-01-20 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of Active Site Inhibitors of the Human Farnesyl Pyrophosphate Synthase: Apoptosis and Inhibition of ERK Phosphorylation in Multiple Myeloma Cells.
J.Med.Chem., 55, 2012
|
|
7SVU
 
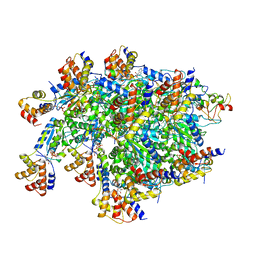 | | TnsBctd-TnsC-TniQ complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (28-MER), DNA (29-MER), ... | | Authors: | Park, J, Tsai, A.W.T, Kellogg, E.H. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the holo CRISPR RNA-guided transposon integration complex
Nature, 613, 2023
|
|
5TUK
 
 | | Crystal structure of tetracycline destructase Tet(51) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Tetracycline destructase Tet(51) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUF
 
 | | Crystal structure of tetracycline destructase Tet(50) in complex with anhydrotetracycline | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
6JCG
 
 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
5TUI
 
 | | Crystal structure of tetracycline destructase Tet(50) in complex with chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5TUE
 
 | | Crystal structure of tetracycline destructase Tet(50) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tetracycline destructase Tet(50) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
6AHG
 
 | | Trimeric structure of concanavalin A from Canavalia ensiformis | | Descriptor: | CADMIUM ION, CALCIUM ION, Concanavalin-A,Concanavalin-A | | Authors: | Park, J.H, Kim, D.S, Park, Y.R, Lee, S.J. | | Deposit date: | 2018-08-18 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Cadmium-substituted concanavalin A and its trimeric complexation
J. Microbiol. Biotechnol., 28(12), 2018
|
|
5TUL
 
 | |
5TUM
 
 | | Crystal structure of tetracycline destructase Tet(56) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tetracycline destructase Tet(56) | | Authors: | Park, J, Tolia, N.H. | | Deposit date: | 2016-11-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Plasticity, dynamics, and inhibition of emerging tetracycline resistance enzymes.
Nat. Chem. Biol., 13, 2017
|
|
6IQ6
 
 | | Crystal structure of GAPDH | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Park, J.B, Park, H.Y. | | Deposit date: | 2018-11-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Study of Monomethyl Fumarate-Bound Human GAPDH.
Mol.Cells, 42, 2019
|
|
8UDK
 
 | | Human Mitochondrial DNA Polymerase gamma R853A Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (24-MER), DNA (28-MER), ... | | Authors: | Park, J, Herrmann, G.K, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
8UDL
 
 | | Human Mitochondrial DNA Polymerase Gamma Binary Complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*CP*GP*AP*CP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*TP*AP*C)-3'), DNA (5'-D(P*AP*GP*GP*TP*AP*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*T)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Park, J, Yin, Y.W. | | Deposit date: | 2023-09-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | An interaction network in the polymerase active site is a prerequisite for Watson-Crick base pairing in Pol gamma.
Sci Adv, 10, 2024
|
|
4GO6
 
 | | Crystal structure of HCF-1 self-association sequence 1 | | Descriptor: | HCF C-terminal chain 1, HCF N-terminal chain 1, SULFATE ION | | Authors: | Park, J, Lammers, F, Herr, W, Song, J. | | Deposit date: | 2012-08-18 | | Release date: | 2012-10-17 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | HCF-1 self-association via an interdigitated Fn3 structure facilitates transcriptional regulatory complex formation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4XQR
 
 | |
5JA0
 
 | | Crystal structure of human FPPS with allosterically bound FPP | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Zielinski, M, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2016-04-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product.
Nat Commun, 8, 2017
|
|
4XQT
 
 | |
4XQS
 
 | |
